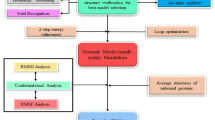
Abstract
Cold shock proteins (CSPs) are induced during abiotic stress, triggered by very low temperatures. These act as “RNA chaperones”, attaching to RNA through their nucleic acid binding domain, to prevent formation of stable, secondary RNA structures, thus initiating translation which is mainly hampered during these aberrant conditions. Computational tools and techniques, such as molecular modelling, energy refinement, normal mode simulation, molecular docking, electrostatic surface potential study, stability and conformational analysis were operated to determine the more stable CSP from the two mesophilic bacteria. The study revealed that cold shock protein from E.coli was more stable than B.subtilis CSP. This was evident from the increased conformational deviations and atomic fluctuations in RMSD and RMSF values during the course of simulation in case of B.subtilis CSP. Interaction study of the CSP-RNA complexes exposed E.coli CSP containing high propensity residues in comparison to B.subtilis. Moreover, stability analysis revealed E.coli to have higher biding affinity and higher SASA energy difference in bound form compared to B.subtilis. E.coli CSP has a more stable complex structure throughout the simulation, with lower deviation in RMSD and RMSF values. It also displays higher binding affinity for ssRNA compared to B.subtilis, thus acting as better “RNA chaperones” during their cold shock response.
Graphical abstract







Similar content being viewed by others
Data Availability
All data generated and studies are available within this and its supplementary article.
Abbreviations
- CSP:
-
Cold shock proteins
- RBM-RNA:
-
Binding Motif
- RMSD:
-
Root Mean Square Deviation
- RMSF:
-
Root Mean Square Fluctuation
- SASA:
-
Solvent-Accessible Surface Area
References
Aier I, Varadwaj PK, Raj U (2016) Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci Rep 6:34984. https://doi.org/10.1038/srep34984
Aziz MF, Caetano-Anollés G (2021) Evolution of networks of protein domain organization. Sci Rep 11:12075. https://doi.org/10.1038/s41598-021-90498-8
Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Research 49:D344–D354. https://doi.org/10.1093/nar/gkaa977
Bornot A, Etchebest C, de Brevern AG (2011) Predicting protein flexibility through the prediction of local structures. Proteins: Struct Function Bioinf 79:839–852. https://doi.org/10.1002/prot.22922
Bowie JU, Ltcy R, Eisenberg D (2014) A method to identify protein sequences that fold into a known Three-Dimensional stucture. Science 253 164 – 70. https://doi.org/10.1126/science.1853201
Cardoza E, Singh H (2022 Feb) Involvement of CspC in response to diverse environmental stressors in Escherichia coli. J Appl Microbiol 132(2):785–801. https://doi.org/10.1111/jam.15219
CHRIS COLOVOS,TODD O. YEATES (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Dominguez C, Boelens R, Bonvin AMJJ (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125:1731–1737. https://doi.org/10.1021/ja026939x
Durham E, Dorr B, Woetzel N, Staritzbichler R, &Meiler J (2009) Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J Mol Model 15:1093–1108. https://doi.org/10.1007/s00894-009-0454-9
Fang LI, Hou Y (1998) Role of the Cold-Box Region in the 5 untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol 180 90 – 5. https://doi.org/10.1128/JB.180.1.90-95.1998
Faßhauer P, Busche T, Kalinowski J, Mäder U, Poehlein A, Daniel R, Stülke J (2021) Functional Redundancy and Specialization of the Conserved Cold Shock Proteins in Bacillus subtilis. Microorganisms.9, 1434. https://doi.org/10.3390/microorganisms9071434
Fraczkiewicz R, Braun W (1998) Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem 19:319–333. https://doi.org/10.1002/(SICI)1096-987X(199802)19:3%3C319::AID-JCC6%3E3.0.CO;2-W
Graumann P, Wendrich TM, Weber MHW, Marahiel MA (1997) A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures.Molecular Microbiology25, 741 – 56. https://doi.org/10.1046/j.1365-2958.1997.5121878.x
Heinig M, Frishman D (2004) STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32:W500–W502. https://doi.org/10.1093/nar/gkh429
Horn G, Hofweber R, Kremer W, Kalbitzer HR (2007) Structure and function of bacterial cold shock proteins. Cell Mol Life Sci 64:1457–1470. https://doi.org/10.1007/s00018-007-6388-4
Kastritis PL, Bonvin AMJJ (2013) On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J Royal Soc Interface 10:20120835. https://doi.org/10.1098/rsif.2012.0835
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Kim H, Jeong E, Lee SW, Han K (2003) Computational analysis of hydrogen bonds in protein-RNA complexes for interaction patterns. FEBS Lett 552:231–239. https://doi.org/10.1016/S0014-5793(03)00930-X
Kishor PB, K (2019) Bacterial cold shock proteins - the Molecular Chaperones for multiple stress tolerance. Adv Biotechnol Microbiol 12(3). https://doi.org/10.19080/AIBM.2019.12.555837
Klose DP, Wallace BA, Janes RW (2010) 2Struc: the secondary structure server. Bioinformatics 26:2624–2625. https://doi.org/10.1093/bioinformatics/btq480
Krüger DM, Ahmed A, Gohlke H (2012) NMSim web server: Integrated approach for normal mode-based geometric simulations of biologically relevant conformational transitions in proteins. Nucleic Acids Res 40:W310–W316. https://doi.org/10.1093/nar/gks478
Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, Laird-Offringa IA (2006) The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res 34:275–285. https://doi.org/10.1093/nar/gkj436
Li Z, Guo Q, Zhang J, Fu Z, Wang Y, Wang T, Tang J (2021) The RNA-Binding motif protein family in Cancer: friend or foe? Front Oncol 11:757135. https://doi.org/10.3389/fonc.2021.757135
Maiti R, van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 3:W590–W594. https://doi.org/10.1093/nar/gkh477
Martínez L (2015) Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE 10:e0119264. https://doi.org/10.1371/journal.pone.0119264
Mukherjee S, Bahadur RP (2018) An account of solvent accessibility in protein-RNA recognition. Sci Rep 8:10546. https://doi.org/10.1038/s41598-018-28373-2
Nakaminami K, Karlson DT, Imai R (2006) Functional conservation of cold shock domains in bacteria and higher plants. PNAS 103:10122–10127. https://doi.org/10.1073/pnas.0603168103
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Porebski BT, Buckle AM (2016) Consensus protein design. In Protein Engineering. Des Selection 29:245–251. https://doi.org/10.1093/protein/gzw015
Receveur-Bréchot V, Durand D (2012) How Random are intrinsically disordered proteins? A small Angle Scattering Perspective. Curr Protein Pept Sci 13:55–75. https://doi.org/10.2174/138920312799277901
Roman A, Laskowski J, Antoon C, Rullmann, Malcolm W, MacArthur, Robert Kaptein, Janet M, Thornton (1996) AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR*. Journal of Biomolecular NMR.8, 477 – 86. https://doi.org/10.1007/BF00228148
Sachs R, Max KEA, Heinemann U, Balbach J (2012) RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. RNA.18, 65–76. https://doi.org/10.1261/rna.02809212
Schindler T, Graumann PL, Perl D, Ma S, Schmid FX, Marahiel MA (1999) The family of cold shock proteins of Bacillus subtilis: Stability and dynamics in vitro and in vivo. J Biol Chem 274:3407–3413. https://doi.org/10.1074/jbc.274.6.3407
Sinha N, Smith-Gill SJ (2002) Electrostatics in Protein Binding and Function. In Current Protein and Peptide Science.3, 601 – 14. https://doi.org/10.1021/acs.chemrev.7b00305
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wiederstein M, &Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research 35:W407–W410. https://doi.org/10.1093/nar/gkm290
Wu S, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35:3375–3382
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534. https://doi.org/10.1016/j.bpj.2011.10.024
Zhao H, Huang D (2011) Hydrogen bonding penalty upon ligand binding. PLoS ONE. https://doi.org/10.1371/journal.pone.0019923
Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y (2021) Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods. https://doi.org/10.1016/j.crmeth.2021.100014
Acknowledgements
The authors would like to thank, Amity Institute of Biotechnology, Amity University, Kolkata, India, for their cooperation.
Funding
No funding support was received during this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
No animals were harmed during this research work.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The supplementary is provided along with the main article.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Roy, A., Ray, S. Exploring RNA-protein interaction between two mesophilic bacteria: an in silico approach to discern detailed molecular level interaction in cold shock response. Biologia 78, 2205–2218 (2023). https://doi.org/10.1007/s11756-023-01352-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-023-01352-3