Abstract
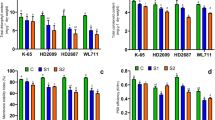
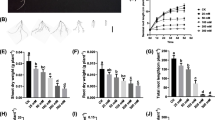
Salt overly sensitive (SOS) pathway genes, SOS1 (plasma membrane Na+/H+ antiporter), SOS2 (CBL interacting protein kinase 24), and SOS3 (calcineurin B like protein 4) are associated with active efflux of toxic sodium ions (Na+) from cytosol and thus confer salinity tolerance in glycophytic plants such as Arabidopsis. The role of SOS pathway genes SOS2 and SOS3 in salinity tolerance of wheat is rarely studied. One-month-old seedlings of three bread wheat genotypes namely, HD 2009, HD2687 and Kharchia 65 were imposed with two levels of salinity stress (100 and 200 mM NaCl) for 30 days duration. Based on the physiological parameters, genotype Kharchia 65 was highly tolerant, HD 2009 was moderately tolerant and HD 2687 was sensitive to salinity stress. Tolerant genotypes accumulated lesser amount of Na+ in root, stem and leaf tissues. Transcript abundance of SOS1, SOS2 and SOS3 genes was significantly higher in salt tolerant genotypes under long-term salinity and correlated with improved sodium exclusion, and higher potassium/sodium (K+/Na+) ratio. Expression levels of genes involved in vacuolar partitioning of Na+, NHX1 (vacuolar Na+/H+ antiporter) and VP1 (Vacuolar pyrophosphatase) were also higher in salt tolerant wheat genotypes under 200 mM NaCl stress. Partial coding sequences of SOS1, SOS2, SOS3, NHX1 and VP1 genes were cloned and sequenced from the above mentioned three wheat genotypes. The results in the present study demonstrated that SOS pathway of ion homeostasis under salinity stress is conserved across species.



Similar content being viewed by others
References
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by over expression of a vacuolar Na+/H+ antiporter in Arabidopsis. Science 285:1256–1258
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S (2011) Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: In planta quantification methods. Plant Cell Environ 34:947–996
Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137:807–818
Feki K, Quintero FJ, Pardo JM, Masmoudi K (2011) Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Mol Biol 76:545–556
Feki K, Quintero FJ, Khoudi H, Leidi EO, Masmoudi K, Pardo JM, Brini F (2014) A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep 33:277–288
Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45:146–159
Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton tranporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96:1480–1485
Gaxiola RA, Li J, Undurraga S, Dang V, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt- tolerant plants result from over expression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98:11444–11449
Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97:3735–3740
Hollington PA (2000) Technological breakthroughs in screening/breeding wheat varieties for salt tolerance. In: Gupta SK, Sharma SK, Tyagi NK (eds) National conference on salinity management in agriculture. Central Soil Salinity Research Institute, Karnal, pp 273–289
Iseki K, Homma K, Shiraiwa T, Jongdee B, Mekwatanakarn P (2014) The effects of cross-tolerance to oxidative stress and drought stress on rice dry matter production under aerobic conditions. Field Crops Res 163:18–23
Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6:275–286
Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Kumar G, Purty RS, Sharma MP, Singla-Pareek SL, Pareek A (2009) Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J Plant Physiol 166:507–520
Lekshmy S, Sairam RK, Kushwaha SR (2013) Effect of long-term salinity stress on growth and nutrient uptake in contrasting wheat genotypes. Ind J Plant Physiol 18(4):344–353
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Martinez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nie WX, Xu L, Yu BJ (2015) A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma 252:127–134
Oh DH, Leidi E, Zhang Q, Hwang SM, Li YZ, Quintero FJ, Jiang XY, D’Urzo MP, Lee SY, Zhao YX (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151:210–222
Olías R, Eljakaoui Z, Li J, Marín- Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32:904–916
Pardo JM, Cubero B, Leidi EO, Quintero FJ (2006) Alkali cation exchangers: roles on cellular homeostasis and stress tolerance. J Exp Bot 57:1181–1199
Plett DC, Moller IS (2010) Na+ transport in glycophytic plants: what we know and would like to know. Plant Cell Environ 33:612–626
Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441
Quintero FJ, Ohta M, Shi HZ, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99:9061–9066
Ramezani A, Ali N, Ali AA, Mahboobeh ZB, Tahereh D, Mahmod E, Hosein A, Esmail E (2013) Quantitative expression analysis of TaSOS1 and TaSOS4 genes in cultivated and wild wheat plants under salt stress. Mol Biotechnol 53:189–197
Sairam RK, Deshmukh PS, Shukla DS (1997) Tolerance to drought and temperature stress in relation to increased antioxidant enzyme activity in wheat. J Agron Crop Sci 178:171–177
Sairam RK, Chandrasekhar V, Srivastava GC (2001) Comparison of hexaploid and tetraploid wheat cultivars in their responses to water stress. Biol Plant 44:89–94
Sairam RK, Rao KV, Srivastava GC (2002) Differential response of wheat genotypes to long-term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci 163:1037–1046
Sanchez-Barrena MJ, Martinez-Ripoll M, Zhu JK, Albert A (2005) The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J Mol Biol 345:1253–1264
Shi H, Zhu JK (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and ABA. Plant Mol Biol 50:543–550
Shi H, Ishitani M, Kim CS, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97:6896–6901
Szabolcs I (1989) Salt-Affected Soils. CRC Press, Boca Raton
Tandon HLS (1995) Estimation of sodium and potassium. Methods of analysis of soils, plants, water and fertilizers. FDCO, New Delhi, pp 62–63
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Weatherley PE (1950) Studies in water relations of cotton plants. I. The field measurement of water deficit in leaves. New Phytol 49:81–87
Xu H, Jiang X, Zhan K, Cheng X, Chen X, Pardo JM, Cui D (2008) Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch Biochem Biophys 473:8–15
Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, Hong XH, Zhu JK, Gong Z (2009) Over expression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2:22–31
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgments
Authors are thankful to the ICAR-Indian Agricultural Research Institute for funding and providing the necessary facilities. Lekshmy Sathee also gratefully acknowledges the Junior and Senior Research Fellowship provided to her by the Council of Scientific and Industrial Research, New Delhi during the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Sowinski.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lekshmy Sathee, Sairam, R.K., Chinnusamy, V. et al. Differential transcript abundance of salt overly sensitive (SOS) pathway genes is a determinant of salinity stress tolerance of wheat. Acta Physiol Plant 37, 169 (2015). https://doi.org/10.1007/s11738-015-1910-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1910-z