Abstract
Radical shifts in reproductive systems result in radical changes in selective pressures acting on reproductive traits. Nematode Caenorhabditis elegans constitutes one of rare model systems where such shifts can be experimentally induced, providing an opportunity for studying the evolution of reproductive phenotypes in real time. Evolutionary history of predominantly selfing reproduction in has led to degeneration of traits involved outcrossing, making it inefficient. Here, we introduced obligatory outcrossing into isogenic lines of C. elegans and allowed replicate populations to evolve under the new reproductive system. We predicted that they should evolve higher outcrossing efficiency, leading to increased fitness relative to unevolved ancestors. To test this prediction, we assayed fitness of both ancestral and evolved outcrossing populations. To control for the potentially confounding effect of adaptation to laboratory conditions, we also assayed populations with wild-type (selfing) reproductive system. In five experimental blocks, we measured competitive fitness of 12 evolved populations (6 outcrossing, 6 selfing) after ca. 95 generations of evolution, along with their respective ancestors. On average, we found that fitness increased by 0.72 SD (± 0.3 CI) in outcrossing and by 0.52 (± 0.35 CI) in selfing populations, suggesting further adaptation to laboratory conditions in both types. Contrary to predictions, fitness increase was not significantly higher in outcrossing populations, suggesting no detectable adaptation to the changed reproductive system. Importantly, the results for individual populations varied strongly between experimental blocks, in some cases even differing in effect direction. This emphasises the importance of experimental replication in avoiding reporting false findings.
Similar content being viewed by others
Introduction
Sexual reproduction sets in motion a number of strong evolutionary forces, shaping a wide range of morphological, physiological and behavioural traits involved in reproductive interactions among individuals (Snook, 2012). Evolutionary shifts in reproductive systems can dramatically alter selective pressures acting on these traits. For example, the evolution of selfing in previously outcrossing taxa is commonly associated with degeneration of traits involved in cross-fertilisation—a phenomenon described as ‘selfing syndrome’ (Cutter, 2008; Shimizu & Tsuchimatsu, 2015). Predominantly, evolutionary consequences of shifts in reproductive systems are studied using comparative methods, inferring on events and processes buried deep in the past. Here, we worked on one of the exceptional systems where such processes can be tracked in real time: the nematode Caenorhabditis elegans. In C. elegans, radical changes in the reproductive system can be introduced experimentally, and subsequent evolution can be tracked for many generations and multiple replicate populations thanks to the nematode’s short life cycle (3–4 days in favourable conditions) and ease of cultivation. We took advantage of these rare opportunities provided by C. elegans in order to investigate populations’ adaptation to altered reproductive mode.
Genus Caenorhabditis includes species reproducing by obligatory outcrossing with separate female and male sexes (dioecious), as well as species consisting mainly of selfing hermaphrodites (which cannot cross-fertilize each other), with occasional males which can fertilize hermaphrodites. C. elegans represents the latter reproductive system, known as androdioecy. Sex is determined by the ratio of X chromosomes to autosomes: XX individuals are hermaphrodites, X0—males (Chandler et al., 2011; Hodgkin, 1987; Hodgkin & Brenner, 1977; Hunter & Wood, 1990). Thus, selfing hermaphrodites produce hermaphrodite offspring, barring very rare cases of sex chromosomes non-disjunction resulting in male progeny, whereas ~ 50% of offspring resulting from male-hermaphrodite outcrossing are males. In wild type populations, the frequency of males is very low and population genomic analyses suggest that the average outcrossing rate is only between 0.01 and 0.001 per generation (Andersen et al., 2012). However, obligatory outcrossing can be induced by introducing mutations which blocks sperm production in hermaphrodites, rendering them incapable of selfing (Anderson et al., 2010) and shifting population sex ratio to ca. 1:1.
Androdioecious Caenorhabditis species, including C. elegans, show numerous symptoms of selfing syndrome. C. elegans males are much less effective at mating than males of related dioecious species: their copulation attempts frequently fail to achieve intromission and when they do succeed, the average copulation time is an order of magnitude shorter than in dioecious C. remanei (Chasnov and Chow, 2002). C. elegans hermaphrodites do not produce male-attracting pheromones found in related dioecious species, tend to move away from males attempting to copulate and can even eject the male’s sperm if they do become inseminated while still having their own sperm available (Kleemann & Basolo, 2007). In populations where male frequency is experimentally elevated and then allowed to evolve, it has been observed to decline exponentially (Stewart & Phillips, 2002), which further supports the conclusion that outcrossing is inefficient in this species. All these symptoms have likely evolved due to relaxed selection for the maintenance of outcrossing-related traits, due to the fact that hermaphrodites do not need mating to reproduce whereas males have become so rare that there is little opportunity for selection to act on their phenotypes. Additionally, they may be driven by positive selection on selfing in hermaphrodites (Cutter, 2008).
The availability of genetic tools to manipulate the reproductive system enables studying the evolution of reproductive phenotypes in real time. In a system where a history of selfing has led to degeneration of traits involved in mating and cross-fertilization, a shift to obligatory outcrossing should re-introduce strong positive selection on such traits, resulting in their rapid evolution. Indeed, some support for this expectation has already been found in experimental evolution studies using C. elegans as a model. For example, Palopoli et al. (2015) found that after 60 generations of evolution under obligatory outcrossing, C. elegans males increased the duration of copulation four times. Previous studies investigating the evolution of reproductive phenotypes in C. elegans under obligatory outcrossing were performed on populations with relatively high initial standing genetic variation. Such populations had to be specially created by crossing several genetically distinct wild isolates, because extant C. elegans populations, both laboratory and natural, are characterised by extremely low genetic variation due to both the primarily selfing mode of reproduction and the fact that new populations are usually founded by few individuals (Richaud et al., 2018).
In this study, we investigated evolution after introducing obligatory outcrossing into C. elegans populations which were initially homogeneous, as is typical in this species. We predicted that adaptation to the novel reproductive system should lead, over generations, to the increase in population-level fitness. We tested this by comparing initially isogenic, obligatorily outcrossing populations before and after 97 generations of evolution under the new reproductive system. We took advantage of the ability of the nematodes to survive freezing, enabling direct comparisons between ancestral and evolved populations in the same experiment. We measured population-level fitness in competitive assays, where both ancestral and evolved populations have competed against a standard marker strain. In order to control for a potential confounding effect of adaptation to the laboratory conditions in which the populations were maintained during experimental evolution, an analogous comparison was performed for populations with wild type (androdioecious) reproductive system, which had been maintained for a similar number of generations (94) in the same laboratory conditions. Since both wild type and outcrossing ancestral lines were derived from lab-adapted N2 strain of C. elegans, we predicted that no further laboratory adaptation should occur in the evolving populations and therefore, that the increase in fitness should be seen only in the outcrossing populations, resulting from adaptation to the novel reproductive system.
In the light of the reliability crisis plaguing modern science (Ioannidis, 2005), replicating experiments is indicated as one of the crucial but often neglected steps to achieve higher reliability of the results and avoid drawing false conclusions (Ioannidis, 2005; Moonesinghe et al., 2007). To this end, five experimental blocks were performed to achieve repeated observations, with every evolved population tested at least in two of them.
Materials and Methods
Experimental Design: Overview
For a detailed description of all procedures, see “Experimental Design: Technical Details”.
Focal Populations
The populations described here are part of a larger scale, ongoing experimental evolution project.
Obligatory outcrossing populations (henceforth referred to as fog populations) have been derived by introgression of fog-2(q71) mutation from JK574 strain on N2 genetic background (for a detailed description of introgression, see Plesnar-Bielak et al., 2017). This mutation blocks sperm production in hermaphrodites, transforming them into functional females, while male spermatogenesis remains unaffected (Schedl & Kimble, 1988). The reproductive system is therefore altered from almost solely selfing with very rare males to obligatory outcrossing with approximately 1:1 sex ratio.
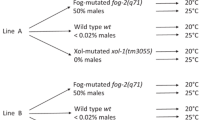
The fog-2(q71) mutation has been introduced independently into replicate isogenic lines with originally wild-type (androdioecious) reproductive system, derived by 20 generations of single-hermaphrodite transfers. This way, from each isoline we obtained a pair of populations differing in the reproductive system: wild-type (wt, without introgressed q71 mutation) and obligatorily outcrossing (fog, with introgressed q71 mutation). Three such pairs were used as ancestral populations for starting experimental evolution (EE). Each ancestral population was split into replicates, some of which were preserved by freezing at − 80 °C for further comparisons, including those reported here (note that the nematodes survive freezing, cf. Introduction), while others were allowed to evolve. While the overall scope of our EE project was broader, the study reported here involved 12 evolutionary populations (6 wt and 6 fog) derived from 2 pairs of ancestral ones (a_wt8 and a_fog8 from isoline 8; a_wt9 and a_fog9 from isoline 9) (Fig. 1). The evolving populations were cultured on 14 cm ø Petri dishes, each generation transferred onto a fresh dish, with census population size maintained at ca. 10,000 individuals. Every 12 ± 3 generations, samples of each population were preserved by freezing (the deviation from 12 was neither systematic nor group-related).
Experimental populations. Two isogenic lines (called isoline 8 and isoline 9) of wild-type C. elegans N2 strain were used to start the experimental populations. For each isoline, the ancestor of wild-type populations (a_wt8 or a_wt9) was in derived directly from the isogenic stock population. The ancestor of fog-2-mutated (‘fog’) populations (a_fog8 and a_fog9) was obtained through introgressing the q71 mutation from C. elegans JK574 strain to the isogenic stock wild-type N2 population. Then, each ancestral population was split into several replicates, which have been evolving independently for the next 94–97 generations
Fitness Assays
Fitness assays compared population-level competitive fitness of evolved populations after 97 (fog populations) or 94 (wt populations) generations of EE with that of their (“unevolved”) ancestors. The assays were performed in 5 experimental blocks, each including evolved and ancestral populations, summarized in Table 1.
The fitness assay workflow (Fig. 2) was identical in all blocks, except for (i) the number of replicate samples per focal population (Table 1) and (ii) the ratio of focal to marker individuals, which differed between e5 and the other 4 blocks (see below). At the beginning of each block, all populations were thawed from samples stored at − 80 °C and allowed 2 generations to recover. We could not assay evolved vs. ancestral populations in direct competition with each other as they are morphologically indistinguishable. Thus, a green fluorescent protein (GFP)-marked strain was used as a standard competitor. Replicate samples from each focal population (evolved or ancestral) were mixed, in standard proportions, with samples of marker strain expressing green fluorescence (GFP). The focal and marker nematodes were allowed to compete for one generation, and the proportions of non-GFP offspring in each sample were scored as fitness measures for the focal populations.
Fitness assay workflow. Only one experimental population is shown for clarity. On Day 1, mixed populations containing a set proportion of GFP and focal L1/L2 stadium larvae were created. After four days, the plate containing both parental animals and their L1/L2 offspring was washed, filtering the adults out. Sample containing larvae was then used to take microscope pictures, from which number of GFP and focal larvae was counted automatically and used to calculate fitness, defined as the ratio of focal larvae to focal plus GFP larvae in the sample
\(fitness = \frac{Nfocal\,larvae}{{Nfocal\,larvae\, +\, NGFP\,larvae}}\).
As the mutation causing GFP expression is dominant, offspring origin in mixed populations can be determined visually: pure fog or wt offspring does not express fluorescence, GFP-GFP as well as mixed GFP-nonGFP offspring express pharyngeal fluorescence. Offspring from the cross of non-GFP male and GFP hermaphrodite is therefore counted as GFP larvae but this should have marginal impact on the results, as most of GFP hermaphrodites offspring is produced via self-fertilization. The opposite effect (offspring produced by GFP male and non-GFP female/hermaphrodite) is also marginal because of GFP males being extremely rare. Similar method was implemented by (Teotonio et al., 2012).
Experimental Design: Technical Details
Experimental Evolution Procedures
During experimental evolution, populations were cultured on 14 cm ø Petri dishes filled with Nematode Growth Medium (NGM), covered with Escherichia coli OP50 strain as a food source, which is a standard procedure (Corsi et al., 2015). The populations were kept in 20 °C incubators and transferred to fresh dishes every ca. four days, which corresponds to one generation time in this temperature. Transfers were performed using filters with 15 µm eyelets, which only let early larvae through (L1–L2 developmental stage) so that a population can be synchronized. During each transfer, each population plate was washed with 4 ml of S Basal solution (Stiernagle, 2006) and the liquid with suspended worms was transferred to a filter positioned on a test tube. The filtered suspension of L1–L2 larvae is vortexed to achieve uniform distribution of larvae in the liquid and the number of transferred animals is counted in 2–3 drops of 1 µl each. Based on this count, the volume of liquid required to transfer 10,000 individuals is estimated and transferred to a fresh plate with bacteria. Every ~ 12 generations samples of each population were frozen to enable further assays of phenotypes from different generations: thanks to the ability of the nematodes to survive freezing, a population can be thawed and propagated further even after long-term freezing. As a rule, the banking was performed every 12 generations; however, for logistic reasons this interval may sometimes vary by a couple of generations.
Marker Strain
GFP-marked animals used for fitness assays were taken from frozen stock populations stored in our laboratory, which had been created by introgression of mls11 transgene from the strain PD4792 on the N2 genetic background. Reproductive system in this populations is, as in the wild type, almost exclusively selfing. These animals exhibit pharyngeal fluorescence from embryos to adults; fluorescence stops immediately after killing the animals with ethanol (personal observation) so selective counting of GFP and non-GFP larvae has to be performed on living animals.
Fitness Assay Procedures
Focal populations (fog and wt, ancestral and evolved) were in all cases thawed and acclimated as follows: cryovial content for each of the populations frozen in − 80 °C was thawed and placed on a 14 cm ø Petri dish with NGM + OP50; nematodes were allowed to complete two generations to recover from freezing before being used in the fitness assays.
To establish the mixed populations for fitness assays (Fig. 2) we placed ~ 1500 focal L1/L2 stadium larvae (fog or wt, evolved or ancestral) and ~ 500 GFP larvae (in blocks 1–4) or ~ 900 focal and ~ 1100 GFP larvae (in block 5) on a 6 cm ø Petri dish. In each case, the resulting density was ~ 71 individuals per cm2 (~ 2000 on 6 cm ø dish), very close to the average population density during experimental evolution (~ 10,000 larvae on 14 cm ø dish = ~ 65/cm2). Mixed populations were kept in standard laboratory conditions until next generation larvae appeared (four days later). Then, dishes were washed with 1 ml S-Basal solution; the liquid was placed on filters which let only small larvae come through. The filtered liquid was put to Eppendorf tubes. After animals sedimented, 5 µl drop was taken from the bottom of the tube (to capture as many animals as possible), placed on a microscope slide and delicately covered with a cover slip. Ten non-overlapping pictures per slide were then taken under magnification ×40 in the Nikon Eclipse 80i microscope with BV-1A filter combination (435/10 nm excitation filter, 470 nm barrier filter, dichromatic mirror value 455 nm), equipped with Nikon Digital Sight DS-U3 camera connected to a computer with NIS-Elements software. Pictures have been analysed using a software based on machine learning, which was previously taught to recognize non-GFP from GFP animals, based on a set of pictures with manually assigned individuals (Palka et al., 2020). The algorithm analyses pictures containing both types of animals by finding nematodes and recognizing them as fluorescent or non-fluorescent. The programme returns marked objects recognized as fluorescent or non-fluorescent nematodes superimposed on an original picture (Fig. 3) as well as a file with a number of animals from each category. The proportion of non-fluorescent larvae in each sample, used as a fitness proxy, was calculated based on data summed from all ten pictures taken from the sample:
Nematode image from fitness assay processed by algorithm classifying them as fluorescent (marked as yellow) or non-fluorescent (markred as green). For details see Palka et al. (2020)
Data Analysis
Data analysis was performed in several steps.
First, we analysed the data separately for each evolved population × block combination. For each evolved population, we ran separate linear models with function lm in R package stats (R Core Team, 2018) for all blocks in which it was assayed, in each block comparing its fitness score to that of its ancestor. To evaluate the magnitude of these differences, in each case we also calculated a relative fitness score of the evolved population (as the percentage of the ancestor’s score) and an effect size (d, known also as Hedges’ d or standardized mean difference (Hedges & Olkin, 1985; Koricheva et al., 2013) with associated confidence intervals. Effect sizes are calculated in standard deviation units (SD) using the formula presented below (1). Thus, biologically, an effect size estimates the change in competitive fitness between an evolved population and its ancestor, measured in the units of phenotypic standard deviation.
where \(\overline{{X_{e} }} - \overline{{X_{c} }}\) is the difference between mean fitness scores of the evolved population and its ancestor, estimated in linear model (“lm estimate” in Online Resource 1; calculated in the model lm(fitness ~ population)), s is pooled standard deviation (2) and J is correction coefficient for small sample size (3):
\(N_{e}\), \(N_{c}\) and \(s_{e}^{2}\), \(s_{c}^{2}\) are numbers of replicate samples and fitness variances in the experimental block of the evolved and ancestral population, respectively. 95% confidence intervals for the effect size were found using the formula (4):
where t is the t value for two-tailed t-test for p = 0.05 and df = Ne + Nc – 2, and \(se_{d}\) is the standard error:
Secondly, we ran meta-analyses to calculate a mean effect size \(\left( \mu \right)\) and its confidence intervals for each experimental population across all experimental blocks in which it was assayed, taking into account differences in number of replicates between blocks:
where d calculated for each population and experimental block was weighted by the inverse of its sampling variance:
95% confidence intervals for mean effect size were found using the formula (10):
where se is calculated with formula (11).
Thirdly, we performed a mixed-effects meta-analysis to assess the overall impact of reproductive type on the evolution of fitness (measured by the difference in fitness scores between evolved and ancestral populations). The analysis was performed with function rma.mv in R package metaphor (Viechtbauer, 2010). This function accounts for non-independence in the observed effects which comes from using the same populations in repeated experimental blocks. In the model (12), effect sizes for each population and experimental block (d, Fig. 6) were used as response variable, variances of d \((V_{d} )\) were used to calculate weights, reproductive type (fog vs. wt) and isoline (Iz8 vs. Iz9) were included as fixed predictors (moderators) and population code was included as random predictor. Since the effect of isoline turned out to be very small and non-significant (Δd = 0.0216 p = 0.935), we removed it from the final model (13), which therefore included reproductive type as the only moderator variable.
Results
Population fitness scores, as well as the results of evolved vs. ancestral population comparisons, varied strongly between experimental blocks. See Figs. 4, 5, and 6 for: distributions of fitness scores for all ancestral and evolved populations in each block (Fig. 4), relative mean fitness scores for all evolved populations (where the ancestor’s mean score is taken as 100%) in each block (Fig. 5), and standardized mean differences (d, ± CI) in fitness scores between each evolved population and its ancestor for each block (Fig. 6). All the estimates are summarized in Online Resource 1; raw data are available in the Figshare repository.
Results of the fitness experiment. Results for populations representing the same reproductive type and isoline are presented in one plot; ancestral populations for are shown with grey fill. All observations are shown as points. Boxes show median and 1st-3rd quartiles, whiskers reach to smallest/largest values within 1.5 of the interquartile range. Asterisks (*) indicate statistically significant difference from ancestor’s fitness (p < 0.05)
Effect sizes for all populations accross all experimental blocks. Numbers on the bars indicate number of replicates of the evolving populations in each experimental block. Number of replicates of the ancestral population might dibffer by ± 1 (Table 1)
Fog Populations
In block 1, fitness scores were significantly higher, relative to the ancestors, in five out of six evolved populations analysed (relative fitness scores: 115–118%, p: 0.003–0.046), and the sixth one showed a nonsignificant trend in the same direction (relative fitness score: 110%, p = 0.126). However, this consistent fitness increase was not confirmed in subsequent experimental blocks (see Figs. 5 and 6). In fact, only a single population (K54 from isoline 8) showed consistently positive (relative fitness scores: 112–123%) although statistically non-significant (p: 0.056–0.274) trends in all following blocks. In other cases, the relative fitness scores varied considerably among blocks, including values < 100% (i.e. lower than in ancestors) for populations K25, K46, and K21. Most strikingly, in two populations (K46 and K21, cf. Fig. 5 and Online Resource 1), relative fitness scores varied from significantly > 100% to significantly < 100%, depending on the block.
Population-level meta-analyses showed that effect sizes were positive for all evolved fog populations, ranging from 0.21 (in K46) to 1.21 (in K54). These net effect sizes assess each evolved population’s divergence from its ancestor in terms of mean fitness, based on data from all blocks in which a given population was assayed, and measured in units of pooled standard deviations (SD) which, in turn, estimate the level of within-population variation in fitness in both the evolved and ancestral populations. Thus, the effect size estimates suggest that mean fitness in fog populations increased by 0.21–1.21 SD (depending on population) over 97 generations of EE. However, the confidence intervals of these estimates were fairly broad and included 0 for all populations except K54 (Fig. 7 and Online Resource 1). Thus, K54 is the only population in which we found conclusive evidence for the evolution of increased fitness.
When data for all populations were summarized in the mixed-effects meta-analysis, we found the mean effect size for all fog populations to be 0.72 ± 0.3 (CI) (Fig. 8) indicating that on average, the populations did increase in fitness relative to their ancestors, even though at the population level this trend was only detectable for K54.
Wt Populations
In wild-type populations, fitness scores also showed high variation across experimental blocks. No population showed a relative fitness score consistently exceeding 100% across all blocks in which it was included. For one population (K37), relative fitness scores varied from 89% (difference from the ancestor not significant statistically; p = 0.373) in one block to 115% (p = 0.079) and 116% (p = 0.028) in two others. Three populations showed a pattern of scoring significantly higher than ancestors in one of the blocks, but non-distinctly from them in the others (K57: relative fitness scores 99%, 134%, 105% with p values 0.899, 0.002, 0.518, respectively; K03: relative fitness scores 97% and 126% with p 0.832 and 0.001, respectively; K43: relative fitness scores 101% and 118% with p 0.922 and 0.040, respectively). One population did not show a significant divergence from the ancestor in either of the blocks it was included in (K24, relative fitness scores: 95% and 105%, p 0.731 and 0.544, respectively). Finally, one population (K16), had fitness score significantly lower than its ancestor (66%, p = 0.021) in one block but slightly and non-significantly higher in two others (110% and 107%, p: 0.184 and 0.324, respectively).
Population-level meta-analyses showed that net effect sizes were positive for all evolved wt populations, ranging from 0.10 (in K24) to 1.02 (in K03); however, these estimates were associated with broad confidence intervals which included 0 for all except one population (K37, mean d = 0.75 ± 0.73 CI) (Fig. 7 and Online Resource 1). It is important to note, however, that two populations (K43 and K03) had higher mean effect sizes than K37 (0.79 and 1.02, respectively), yet their confidence intervals overlapped 0 because they were broader than in K37 (Fig. 7 and Online Resource 1). These differences in precision of mean effect size estimates were partly due to differences in replication: K43 and K03 were only assayed in two experimental blocks (e2 and e3) with small per-population sample sizes (3 and 4, respectively), whereas K37 was assayed in three blocks: e2, e3 as well as e4 (with per-population sample sizes of 9 in the latter) (Table 1).
The mixed-effects meta-analysis summarizing data for all populations revealed that the overall mean effect size for wt populations was 0.52 ± 0.35 CI. Thus, similarly as in fog, we also found evidence for mean fitness increase in wt populations over 94 generations of EE, although at the population level this trend was only statistically detectable in one population (K37).
Fog vs. Wt
The overall mean effect size for wt populations (0.52 ± 0.35 CI) was lower than that for fog populations (0.72 ± 0.3 CI); however, each reproductive system’s confidence intervals overlapped the other’s mean estimate. Thus, our results indicate that populations of both reproductive systems evolved increased population-level fitness (relative to their ancestors) over ca. 95 generations of experimental evolution, although at the level of individual populations this effect was only statistically detectable in two cases: K54 (fog) and K37 (wt).
Discussion
Rapid Evolution from No Initial Genetic Variation
As outlined in the Introduction, we expected that inducing obligatory outcrossing in C. elegans populations would incur strong selection on outcrossing-related traits– and that adaptation to the novel reproductive system would manifest as increased population-level fitness (measured as competitive fitness with a marker strain). Indeed, our results indicate that over ca. 95 generations of evolution, fitness did increase in obligatorily outcrossing (fog) lines, although at the level of individual populations this effect was statistically detectable in only one of them (cf. Figs. 7 and 8). However, a very similar pattern observed in populations evolving under wild type reproductive system (wt) suggests that fitness increase in both fog and wt lines may reflect their adaptation to laboratory conditions, rather than the fog populations’ adaptation to the novel reproductive system. Thus, we did not find evidence that adaptation to outcrossing had occurred in our fog populations—at least not in ways and/or at the level detectable by the assays we applied.
Instead, our results suggest laboratory adaptation occurring in populations of both reproductive systems. This was surprizing, because the C. elegans strain used in the experiment (N2) had already undergone long-term and extensive adaptation to laboratory conditions (Sterken et al., 2015). Nevertheless, adaptation to the particular conditions characterizing our lab and our EE setup could have been happening—and fitness gains observed in wt populations indicate just so. Although not in the way we predicted, our results suggest that rapid and repeatable adaptation starting from no initial genetic variation is occurring in our EE lines. While the effect was statistically detectable in only two cases when individual populations were analysed separately, the mixed-effects meta-analysis summarizing data for all populations revealed a significant upward shift in fitness between ancestors and their descendants after ca. 95 generations.
Rapid evolution from new mutations is possible under strong selection, provided that suitable genetic variants arise. For example, (Pascoal et al., 2014) observed rapid convergent evolution of a new trait—altered wing morphology in wild crickets on two neighbouring Hawaiian islands, as a response to selective pressure imposed by a parasitoid. On each of the two islands, a different genetic change underlying this phenotype has arisen and spread independently within ca. 20 generations (Zuk et al., 2006). Another example of fast and repeatable evolution from new genetic variants comes from the studies of resistance to quinone outside inhibiting (QoI) fungicide in Mycosphaerella graminicola, a pest of wheat. In four distant locations in Europe, the same resistance-conferring mutation appeared independently and spread within two years (Torriani et al., 2009). In C. elegans, (Denver et al., 2010) found a fast adaptive response in five independent populations initiated from a low-fitness, isogenic progenitor line and allowed to evolve independently at population size of ca. 1000 individuals for 60 generations. All five populations rapidly evolved increased fitness and subsequent genomic analysis showed that this increase was associated with very fast (within 10–20 generations) fixation of new mutations. Other examples come from studies on artificial selection on new mutations. In C. elegans (Azevedo et al., 2002) observed that in two isogenic lines selected in the opposite directions of body size change, after 48 generations the body size decreased or increased by 35 and 8%, respectively. In Drosophila melanogaster, artificial selection experiments on bristle number also show that within 20–40 generations of evolution of initially isogenic populations, significant differences can occur (Caballero et al., 1991; Mackay et al., 1994; Merchante et al., 1995). Even in a mammalian species, Mus musculus, new mutations can be a source of a detectable response after 25–50 generations of selection on body size (Keightley, 1998; Keightley & Hill, 1992), although it has been shown that in this case also epistatic interactions between these mutations can play an important role in adaptation (Casellas et al., 2014).
Outcrossing alters associations among alleles within populations’ genomes. This effect may increase the efficiency of selection on individual alleles, facilitating adaptation (Otto, 2009; McDonald et al., 2016). In the light of the evidence for laboratory adaptation found in our study, this would still predict higher fitness gains in outcrossing populations. However, we did not observe such effect. Longer-term evolution would be needed to check whether it would arise with more generations available for genetic shuffling.
Notes of Caution and a Case Study for the Repeatability Crisis in (Evolutionary) Biology
Although this study was designed in order to address specific biological questions, we feel that some of the main conclusions concern very important methodological issues which, despite being discussed in scientific literature for at least a couple of decades, are still extremely rarely taken into account in actual research, at least in the evolutionary biology community. The most striking finding in this study was not directly related with our biological questions, but concerned the unreliability of the results obtained in single experimental blocks: for the majority of the evolved populations assayed, radically different conclusions regarding their fitness divergence from ancestors would have been drawn based on data from single blocks, depending on which block would be considered (Figs. 5, 6). This striking level of non-replication has forced us to delve into literature concerning the repeatability/reliability crisis. This crisis has been recognized in biological and social sciences as an extremely serious problem: many authors convincingly suggest that in fact, most published research findings are false (e.g. Ioannidis, 2005; Moonesinghe et al., 2007). Precisely due to the generally high risks of producing false findings, replicating experiments is a key tool which should be used whenever possible to verify research conclusions (Moonesinghe et al., 2007; Parker 2013).
Conclusions
In both reproductive types significant fitness gain after 94/97 generations was detected, which indicates further adaptation to laboratory conditions. In contrary to predictions, fitness increase was not significantly higher in fog than in the wild-type populations. Another striking finding was the degree of variability among experimental blocks, emphasizing the importance of experiment replication.
Data Availability
The datasets generated and analysed during the current study are available in the Figshare repository, https://doi.org/10.6084/m9.figshare.17429699.v1.
References
Andersen, E. C., et al. (2012). Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nature Genetics, 44(3), 285–290. https://doi.org/10.1038/ng.1050
Anderson, J. L., Morran, L. T., & Phillips, P. C. (2010). Outcrossing and the maintenance of males within C. elegans populations. Journal of Heredity. https://doi.org/10.1093/jhered/esq003
Azevedo, R. B. R., et al. (2002). Spontaneous mutational variation for body size in Caenorhabditis elegans. Genetics Society of America, 162, 755–765.
C Mackay, T. F., et al. (1994). Polygenic mutation in Drosophila melanogaster: Estimates from response to selection of inbred strains. Genetics, 136, 937–951.
Caballero, A., Toro, M. A., & Lopez-Fanjul, C. (1991). The response to artificial selection from new mutations in Drosophila melanogaster. Genetics, 127, 89–102.
Casellas, J., Gianola, D., & Medrano, J. F. (2014). Bayesian analysis of additive epistasis arising from new mutations in mice. Genetics Research, 96, e008. https://doi.org/10.1017/S001667231400010X
Chandler, C. H., et al. (2011). Experimental evolution of the Caenorhabditis elegans sex determination pathway. Evolution, 66(1995), 82–93. https://doi.org/10.5061/dryad.cg872
Corsi, A. K., Wightman, B., & Chalfie, M. (2015). A Transparent window into biology: A primer on Caenorhabditis elegans. WormBook. https://doi.org/10.1895/wormbook.1.177.1
Cutter, A. D. (2008). Reproductive evolution: Symptom of a selfing syndrome. Current Biology, 18(22), 1056–1058. https://doi.org/10.1016/j.cub.2008.09.008
Denver, D. R., et al. (2010). Selective sweeps and parallel mutation in the adaptive recovery from deleterious mutation in Caenorhabditis elegans. Genome Research, 20(12), 1663–1671. https://doi.org/10.1101/gr.108191.110
Hedges, L., & Olkin, I. (1985). Statistical methods for meta-analysis. Orlando: Academic Press.
Hodgkin, J. (1987). Sex determination and dosage compensation in Caenorhabditis elegans. Annual Review of Genetics, 21, 133–154.
Hodgkin, J. A., & Brenner, S. (1977). Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics, 86, 275–287.
Hunter, C. P., & Wood, W. B. (1990). The tra-1 gene determines sexual phenotype cell-autonomously in C. elegans. Cell, 63(6), 1193–1204. https://doi.org/10.1016/0092-8674(90)90415-B
Ioannidis, J. P. A. (2005). Why most published research findings are false. Getting to Good: Research Integrity in the Biomedical Sciences, 2(8), 2–8. https://doi.org/10.1371/journal.pmed.0020124
Keightley, P. D. (1998). Genetic basis of response to 50 generations of selection on body weight in inbred mice. Genetics, 148, 1931–1939.
Keightley, P. D., & Hill, W. G. (1992). Quantitative genetic variation in body size of mice from new mutations. Genetics, 131, 693–700.
Kleemann, G. A., & Basolo, A. L. (2007). Facultative decrease in mating resistance in hermaphroditic Caenorhabditis elegans with self-sperm depletion. Animal Behaviour, 74(5), 1339–1347. https://doi.org/10.1016/j.anbehav.2007.02.031
Koricheva, J., Gurevitch, J., & Mengersen, K. (2013). Handbook of meta-analysis in ecology and evolution. Princeton University Press.
McDonald, M. J., Rice, D. P., & Desai, M. M. (2016). Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. https://doi.org/10.1038/nature17143
Merchante, M., Caballero, A., & Lopez-Fanjul, C. (1995). Response to selection from new mutation and effective size of partially inbred populations. II. Experiments with Drosophila melanogaster. Genetics Research, 66, 227–240. https://doi.org/10.1017/S0016672300034674
Moonesinghe, R., Khoury, M. J., & Janssens, A. C. J. W. (2007). Most published research findings are false—but a little replication goes a long way. PLoS Medicine, 4(2), e28. https://doi.org/10.1371/journal.pmed.0040028
Otto, S. P. (2009). The evolutionary enigma of sex. The American Naturalist, 174(S1), S1–S14.
Palka, J. K., et al. (2020). Competitive fitness analysis using convolutional neural network. Journal of Nematology. https://doi.org/10.21307/JOFNEM-2020-108
Palopoli, M. F., et al. (2015). Natural and experimental evolution of sexual conflict within Caenorhabditis nematodes. BMC Evolutionary Biology. https://doi.org/10.1186/s12862-015-0377-2
Pascoal, S., et al. (2014). Rapid convergent evolution in wild crickets. Current Biology, 24, 1369–1374. https://doi.org/10.1016/j.cub.2014.04.053
Plesnar-Bielak, A., et al. (2017). Fitness effects of thermal stress differ between outcrossing and selfing populations in Caenorhabditis elegans. Evolutionary Biology, 44, 356–364. https://doi.org/10.1007/s11692-017-9413-z
R Core Team. (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing.
Richaud, A., et al. (2018). The local coexistence pattern of selfing genotypes in Caenorhabditis elegans natural metapopulations. Genetics, 208, 807–821.
Schedl, T., & Kimble, J. (1988). fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics, 119, 43–61.
Shimizu, K. K., & Tsuchimatsu, T. (2015). Evolution of selfing: Recurrent patterns in molecular adaptation. Annual Review of Ecology Evolution and Systematics, 46, 593–622. https://doi.org/10.1146/annurev-ecolsys-112414-054249
Snook, R. R. (2012). The evolution of anisogamy: A fundamental phenomenon underlying sexual selection | The evolution of anisogamy: A fundamental phenomenon underlying sexual selection. Animal Behaviour, 84(2), 495–496. https://doi.org/10.1016/J.ANBEHAV.2012.05.014
Sterken, M. G., et al. (2015). The laboratory domestication of Caenorhabditis elegans. Trends in Genetics, 31, 224–231. https://doi.org/10.1016/j.tig.2015.02.009
Stewart, A. D., & Phillips, P. C. (2002). Selection and maintenanace of androdioecy in Caenorhabditis elegans. Genetics, 160, 975–982.
Stiernagle, T. (2006). Maintenance of C. elegans. WormBook. https://doi.org/10.1895/wormbook.1.101.1
Teotonio, H., et al. (2012). Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS ONE, 7(4), e35811. https://doi.org/10.1371/journal.pone.0035811
Torriani, S. F. F., et al. (2009). QoI resistance emerged independently at least 4 times in European populations of Mycosphaerella graminicola. Pest Management Science, 65(2), 155–162. https://doi.org/10.1002/ps.1662
Viechtbauer, W. (2010). Conducting meta-analyses in R with the metafor package. Journal of Statistical Software, 36(3), 1–48. https://doi.org/10.18637/jss.v036.i03
Zuk, M., Rotenberry, J. T., & Tinghitella, R. M. (2006). Silent night: Adaptive disappearance of a sexual signal in a parasitized population of field crickets. Biology Letters, 2(4), 521–524. https://doi.org/10.1098/rsbl.2006.0539
Acknowledgements
Conducting the experimental blocks e1 and e2 was financed from the National Science Centre (Poland) Grant 2018/29/N/NZ8/02616 to Weronika Antoł and blocks e3-e5 from the National Science Centre (Poland) Grant 2013/09/B/NZ8/03317 to Zofia M. Prokop
Author information
Authors and Affiliations
Contributions
Conceptualization: ZMP, MKL; Methodology: all authors; Investigation: WA, JKP, AB, KS, PK; Formal analysis: WA, ZMP, JKP; Visualisation: WA; Writing—original draft preparation: WA, ZMP; Writing—review and editing: all authors; Funding acquisition: ZMP, WA; Supervision: ZMP.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The experiments comply with the current laws of the country in which they were performed (Poland). No approval of research ethics committees was required to accomplish the goals of this study because experimental work was conducted with an unregulated invertebrate species. The research institution possesses valid permits to use genetically modified organisms (GFP-marked C. elegans strain).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Antoł, W., Palka, J.K., Błażejowska, A. et al. Evolution of Reproductive Efficiency in Caenorhabditis elegans Under Obligatory Outcrossing. Evol Biol 49, 314–326 (2022). https://doi.org/10.1007/s11692-022-09572-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-022-09572-2