Abstract
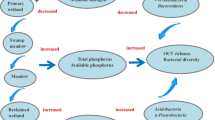
Amplicon sequencing of functional genes is a powerful technique to explore the diversity and abundance of microbes involved in biogeochemical processes. One such key process, denitrification, is of particular importance because it can transform nitrate (NO3-) to N2 gas that is released to the atmosphere. In nitrogen limited alpine wetlands, assessing bacterial denitrification under the stress of wetland desertification is fundamental to understand nutrients, especially nitrogen cycling in alpine wetlands, and thus imperative for the maintenance of healthy alpine wetland ecosystems. We applied amplicon sequencing of the nirS gene to analyze the response of denitrifying bacterial community to alpine wetland desertification in Zoige, China. Raw reads were processed for quality, translated with frameshift correction, and a total of 95,316 nirS gene sequences were used for rarefaction analysis, and 1011 OTUs were detected and used in downstream analysis. Compared to the pristine swamp soil, edaphic parameters including water content, organic carbon, total nitrogen, total phosphorous, available nitrogen, available phosphorous and potential denitrification rate were significantly decreased in the moderately degraded meadow soil and in severely degraded sandy soil. Diversity of the soil nirS-type denitrifying bacteria communities increased along the Zoige wetland desertification, and Proteobacteria and Chloroflexi were the dominant denitrifying bacterial species. Genus Cupriavidus (formerly Wautersia), Azoarcus, Azospira, Thiothrix, and Rhizobiales were significantly (P <0.05) depleted along the wetland desertification succession. Soil available phosphorous was the key determinant of the composition of the nirS gene containing denitrifying bacterial communities. The proportion of depleted taxa increased along the desertification of the Zoige wetland, suggesting that wetland desertification created specific physicochemical conditions that decreased the microhabitats for bacterial denitrifiers and the denitrification related genetic diversity.
Similar content being viewed by others
References
Abell GCJ, Ross DJ, Keane JP, et al. (2013) Nitrifying and denitrifying microbial communities and their relationship to nutrient fluxes and sediment geochemistry in the Derwent Estuary, Tasmania. Aquatic Microbial Ecology 70(1): 63–75. https://doi.org/10.3354/ame01642
Arroyo P, Saenz de Miera LE, Ansola G (2015) Influence of environmental variables on the structure and composition of soil bacterial communities in natural and constructed wetlands. Science of the Total Environment 506–507 (506–507C): 380–390. https://doi.org/10.1016/j.scitotenv.2014.11.039
Báttard E, Rechová T, Vaněk D, et al. (2010) Effect of pH and dissolved organic matter on the abundance of nirK and nirS denitrifiers in spruce forest soil. Biogeochemistry 101(1–3): 123–132. https://doi.org/10.1007/s10533-010-9430-9
Bender SF, Plantenga F, Neftel A, et al. (2014) Symbiotic relationships between soil fungi and plants reduce N2O emissions from soil. The ISME Journal 8: 1336–1345. https://doi.org/10.1038/ismej.2013.224
Bremer C, Braker G, Matthies D, et al. (2007) Impact of plant functional group, plant species and sampling time on the composition of nirK-type denitifier communities in soil. Applied and Environmental Microbiology 73(21): 6876–6884. https://doi.org/10.1128/AEM.01536-07
Caporaso JG, Lauber CL, Walters W A, et al. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal 6(8): 1621–1624. https://doi.org/10.1038/ismej.2012.8
Chmolowska D, Kozak M, Laskowski R. (2016) Soil physicochemical properties and floristic composition of two ecosystems differing in plant diversity: fallows and meadows. Plant and Soil 402: 317–329. https://doi.org/10.1007/s11104-015-2788-7
Cui X, Graf H (2009) Recent land cover changes on the Tibetan Plateau: a review. Climatic Change 94(1–2): 47–61. https://doi.org/10.1007/s10584-009-9556-8
Cytryn E, Minz D, Gelfand I, et al. (2005) Sulfide-oxidizing activity and bacterial community structure in a fluidized bed reactor from a zero-discharge mariculture system. Environmental Science and Technology 39(6): 1802–1810. https://doi.org/10.1021/es0491533
Dambreville C, Séphanie H, Nguyen C, et al. (2006) Structure and activity of the denitrifying community in a maize-cropped field fertilized with composted pig manure or ammonium nitrate. FEMS Microbiology Ecology 56(1): 119–131. https://doi.org/10.1111/j.1574-6941.2006.00064.x
Gao MH, Liu JW, Qiao YL, et al. (2017) Diversity and abundance of the denitrifying microbiota in the sediment of eastern China Marginal Seas and the impact of environmental factors. Microbial Ecology 73(3): 602–615. https://doi.org/10.1007/s00248-016-0906-6
Gu YF, Wang YY, Xiang QJ, et al. (2017) Implications of wetland degradation for the potential denitrifying activity and bacterial populations with nirS genes as found in a succession in Qinghai-Tibet plateau, China. European Journal of Soil Biology 80: 19–26. https://doi.org/10.1016/j.ejsobi.2017.03.005
Gu YF, Bai Y, Xiang QJ, et al. (2018) Degradation shaped bacterial and archaeal communities with predictable taxa and their association patterns in Zoige wetland at Tibet plateau. Scientific Reports 8(1): 3884. https://doi.org/10.1038/s41598-018-21874-0
Hong X, Zhang XJ, Liu BB, et al. (2010) Structural differentiation of bacterial communities in indole-degrading bioreactors under denitrifying and sulfate-reducing conditions. Research in Microbiology 161(8): 687–693. https://doi.org/10.1016/j.resmic.2010.06.010
Huo L, Chen Z, Zou Y, et al. (2013) Effect of Zoige alpine wetland degradation on the density and fractions of soil organic carbon. Ecological engineering 51(1): 287–295. https://doi.org/10.1016/j.ecoleng.2012.12.020
Hwang C, Wu WM, Gentry TJ, et al. (2006) Changes in bacterial community structure correlate with initial operating conditions of a field-scale denitrifying fluidized bed reactor. Applied Microbiology and Biotechnology 71(5): 748–760. https://doi.org/10.1007/s00253-005-0189-1
Jones CM, Hallin S. (2010) Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. The ISME Journal 4(5): 633–641. https://doi.org/10.1038/ismej.2009.152
Kato T, Tang YH, Gu S, et al. (2006) Temperature and biomass influences on inter annual changes in CO2 exchange in an alpine meadow on the Qinghai-Tibetan Plateau. Global Change Biology 12(7): 1285–1298. https://doi.org/10.1111/j.1365-2486.2006.01153.x
Katsuyama C, Nashimoto H, Nagaosa K, et al. (2013) Occurrence and potential activity of denitrifiers and methanogens in groundwater at 140 m depth in Pliocene diatomaceous mudstone of northern Japan. FEMS Microbiology Ecology 86(3):532–543. https://doi.org/10.1111/1574-6941.12179
Kim YM, Cho HU, Lee DS, et al. (2011) Influence of operational parameters on nitrogen removal efficiency and microbial communities in a full-scale activated sludge process. Water research 45(17): 5785–5795. https://doi.org/10.1016/j.watres.2011.08.063
Kumar S, Stecher G, Tamura K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. https://doi.org/10.1093/molbev/msw054
Lee JA, Francis CA. (2017) Deep nirS amplicon sequencing of San Francisco Bay sediments enables prediction of geography and environmental conditions from denitrifying community. Envionmental Microbiology 19(12): 4897–4912. https://doi.org/10.1111/1462-2920.13920
Liang YT, Van Nostrand JD, Deng Y, et al. (2011) Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. The ISME Journal 5(3): 403–413. https://doi.org/10.1038/ismej.2010.142
Lisa JA, Jayakumar A, Ward BB, et al. (2017) nirS-type denitrifying bacterial assemblages respond to environmental conditions of a shallow estuary. Environmental Microbiology Reports 9(6): 766–778. https://doi.org/10.1111/1758-2229.12594
Love MI, Huber W, Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15(12): 550. https://doi.org/10.1186/s13059-014-0550-8
Ludwig W, Strunk O, Westram R, et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Research 32(4): 1363–1371.https://doi.org/10.1093/nar/gkh293
Oksanen J, Blanchet FG, Friendly M, et al. (2018) Package: ‘Vegan’. Community Ecology Package. R package version 2.0±3. https://doi.org/github.com/vegandevs/vegan
Osaka T, Shirotani K, Yoshie S, et al. (2008) Effects of carbon source on denitrification efficiency and microbial community structure in a saline wastewater treatment process. Water Research 42(14): 3709. https://doi.org/10.1016/j.watres.2008.06.007
Piña-Ochoa E, Álvarez-Cobelas M. (2006) Denitrification in aquatic environments: a cross-system analysis. Biogeochemistry 81(1): 111–130. https://doi.org/10.1007/s10533-006-9033-7
R Development Core Team (2010) R: a language and environment for statistical computing. Vienna: The R Foundation for Statistical Computing. https://doi.org/10.1007/978-1-4614-3520-4_41
Rungkitwatananukul P, Nomai S, Hirakata Y, et al. (2016) Microbial community analysis using MiSeq sequencing in a novel configuration fluidized bed reactor for effective denitrification. Bioresource Technology 221: 677–681. https://doi.org/10.1016/j.biortech.2016.09.051
Smith CJ, Nedwell DB, Dong LF, et al. (2007) Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Applied and Environmental Microbiology 73(11): 3612–3622. https://doi.org/10.1002/gepi.10246
Veraart AJ, Dimitrov MR, Schrier-Uijl AP, et al. (2017) Abundance, activity and community structure of denitrifiers in drainage ditches in relation to sediment characteristics, vegetation and land-use. Ecosystems 20(5): 928–943. https://doi.org/10.1007/s10021-016-0083-y
Wang C, Tong C, Chambers LG, et al. (2017) Identifying the salinity thresholds that impact greenhouse gas production in subtropical tidal freshwater marsh soils. Wetland 37(3): 559–571. https://doi.org/10.1007/s13157-017-0890-8
Wang Q, Quensen JF, Fish JA, et al. (2013) Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio 4(5): e00592–13. https://doi.org/10.1128/mBio.00592-13
Wang YY, Lu SE, Xiang QJ. (2017) Responses of N2O reductase gene (nosZ)-denitrifier communities to long-term fertilization follow a depth pattern in calcareous purplish paddy soil. Journal of Integrative Agriculture 16(11): 60345–7. https://doi.org/10.1016/S2095-3119(17)61707-6
Wang Y, Tian H, Huang F, et al. (2017) Time-resolved analysis of a denitrifying bacterial community revealed a core microbiome responsible for the anaerobic degradation of quinoline. Scientific Reports 7(1): 14778. https://doi.org/10.1038/s41598-017-15122-0
Wang Y, Uchida Y, Shimomura Y, et al. (2017) Responses of denitrifying bacterial communities to short-term waterlogging of soils. Scientific Reports 7(1): 803. https://doi.org/10.1038/s41598-017-00953-8
Ward BB (2013) How nitrogen is lost. Science 341(6144): 352–353. https://doi.org/10.1126/science.1240314
Wu LS, Nie YY, Yang ZR, et al. (2016) Responses of soil inhabiting nitrogen-cycling microbial communities to wetland degradation on the Zoige Plateau, China. Journal of Mountain Science 13(22): 2192–2204. https://doi.org/10.1007/s11629-016-4004-5
Yang YD, Hu YG, Wang ZM, et al. (2018) Variations of the nirS-, nirK-, and nosZ-denitrifying bacterial communities in a northern Chinese soil as affected by different long-term irrigation regimes. Environmental Science and Pollution Research 25 (14): 14057–14067. https://doi.org/10.1007/s11356-018-1548-7
Yin C, Fan FL, Song AL, et al. (2015) Denitrification potential under different fertilization regimes is closely coupled with changes in the denitrifying community in a black soil. Applied Microbiology and Biotechnology 99(13): 5719–5729. https://doi.org/10.1007/s00253-015-6461-0
Yu ZH, Liu JJ, Li YS, et al. (2018) Impact of land use, fertilization and seasonal variation on the abundance and diversity of nirS-type denitrifying bacterial communities in a Mollisol in Northeast China. European Journal of Soil Biology 85: 4–11. https://doi.org/10.1016/j.ejsobi.2017.12.001
Yun JL, Zhuang GQ, Ma AZ, et al. (2012) Community structure, abundance, and activity of methanotrophs in the Zoige wetland of the Tibetan plateau. Microbial Ecology 63(4): 835–843. https://doi.org/10.2307/41489227
Zhong Q, Chen H, Liu L, et al. (2017) Water table drawdown shapes the depth-dependent variations in prokaryotic diversity and structure in Zoige peatlands. FEMS Microbiology Ecology 93(6): 049. https://doi.org/10.1093/femsec/fix049
Acknowledgments
The study was supported by the Natural Science Foundation of China (Grant No. 41201256).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Gu, Yf., Liu, T., Bai, Y. et al. Pyrosequencing of nirS gene revealed spatial variation of denitrifying bacterial assemblages in response to wetland desertification at Tibet plateau. J. Mt. Sci. 16, 1121–1132 (2019). https://doi.org/10.1007/s11629-018-5147-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11629-018-5147-3