Abstract
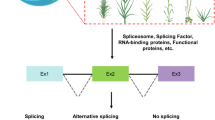
Callus induction in plants is similar to pluripotent stem cell induction in animals and can incite global changes in gene expression. Alternative splicing is considered a key factor underlying functional complexity and function in the induction of pluripotent stem cells in animals. However, the role of alternative splicing in plant callus induction remains unclear. We performed high-throughput RNA sequencing of maize callus induced on a callus induction medium (CIM) at different stages and recorded global alternative splicing changes. At every stage, over 20,000 alternative splicing events were detected, and the numbers slightly increased with callus induction. In total, 648 to 2580 genes were found to exhibit significant alternative splicing changes during callus induction—mainly those that belong to the spliceosome, metabolic pathways, and mRNA surveillance pathways. It was also found that alternative splicing can function alongside transcriptional regulation to contribute to callus induction. Especially, ZmWOX11, whose orthologous gene in Arabidopsis plays a vital role in the first-step cell fate transition of callus induction, was found to exhibit alternative splicing and expression level changes during callus induction. Our results establish a foundation from which researchers can further explore the molecular mechanism of callus induction in maize, especially at alternative splicing level.






Similar content being viewed by others
References
Andorf CM, Cannon EK, Portwood JL, Gardiner JM, Harper LC, Schaeffer ML, Braun BL, Campbell DA, Vinnakota AG, Sribalusu VV (2016) MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res 44:D1195–D1201
Armstrong CL, Romero SJ, Hodges TK (1992) Improved tissue culture response of an elite maize inbred through backcross breeding, and identification of chromosomal regions important for regeneration by RFLP analysis. Theor Appl Genet 84:755–762
Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW (2008) OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cell. Stem Cells 26:3068–3074
Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126:37–47
Cáceres JF, Kornblihtt AR (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18:186–193
Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76:51–74
Cheng S, Tan F, Lu Y, Liu X, Li T, Yuan W, Zhao Y, Zhou DX (2018) WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res 46:2356–2369
Dong C, He F, Berkowitz O, Liu J, Cao P, Tang M, Shi H, Wang W, Li Q, Shen Z, Whelan J, Zheng L (2018) Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice ( Oryza sativa ). Plant Cell 30:2267–2285
Duclercq J, Sangwan NB, Catterou M, Sangwan RS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16:597–606
Fan M, Xu C, Xu K, Hu Y (2012) Lateral organ boundaries domain transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22:1169–1180
Filichkin S, Priest HD, Megraw M, Mockler TC (2015) Alternative splicing in plants: directing traffic at the crossroads of adaptation and environmental stress. Curr Opin Plant Biol 24:125–135
Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC (2010) Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 20:45–58
Foissac S, Sammeth M (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35:W297–W299
Fransz PF, Kieft H, Schel JHN (1990) Cell cycle changes during callus initiation from cultured maize embryos. An autoradiographic study. Acta Bot Neerl 39:65–73
Gallois JL, Nora FR, Mizukami Y, Sablowski R (2004) WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 18:375–380
Ge F, Luo X, Huang X, Zhang Y, He X, Liu M, Lin H, Peng H, Li L, Zhang Z, Pan G, Shen Y (2016) Genome-wide analysis of transcription factors involved in maize embryonic callus formation. Physiol Plant 158:452–462
Gracheva EO, Cordero-Morales JF, Gonzalez-Carcacia JA, Ingolia NT, Manno C, Aranguren CI, Weissman JS, Julius D (2011) Ganglion-specific splicing of TRPV1 underlies infrared sensation in vampire bats. Nature 476:88–91
Guilfoyle TJ (2015) The PB1 domain in auxin pesponse factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27:33–43
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460
Hu B, Zhang G, Liu W, Shi J, Wang H, Qi M, Li J, Qin P, Ruan Y, Huang H, Zhang Y, Xu L (2017) Divergent regeneration-competent cells adopt a common mechanism for callus initiation in angiosperms. Regeneration 4:132–139
Ikeuchi M, Sugimoto K, Iwase A (2013) Plant callus: mechanisms of induction and repression. Plant Cell 25:3159–3173
Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40:77–105
Ishida Y, Hiei Y, Komari T (2007) Agrobacterium-mediated transformation of maize. Nat Protoc 2:1614–1621
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci 111:5427–5432
Kriechbaumer V, Wang P, Hawes C, Abell BM (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation: YUCCA4 and auxin biosynthesis. Plant J 70:292–302
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li SB, OuYang WZ, Hou XJ, Xie LL, Hu CG, Zhang JZ (2015) Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front Plant Sci 6:119
Li SB, Xie ZZ, Hu CG, Zhang JZ (2016) A review of auxin response factors (ARFs) in plants. Front Plant Sci 7:47
Liu H, Ma L, Yang X, Zhang L, Zeng X, Xie S, Peng H, Gao S, Lin H, Pan G, Wu Y, Shen Y (2017) Integrative analysis of DNA methylation, mRNAs, and small RNAs during maize embryo dedifferentiation. BMC Plant Biol 17:105
Liu J, Hu X, Qin P, Prasad K, Hu Y, Xu L (2018a) The WOX11–LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol 59:739–748
Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26:1081–1093
Liu Z, Li J, Wang L, Li Q, Lu Q, Yu Y, Li S, Bai MY, Hu Y, Xiang F (2016) Repression of callus initiation by the miRNA-directed interaction of auxin-cytokinin in Arabidopsis thaliana. Plant J 87:391–402
Liu Z, Qin J, Tian X, Xu S, Wang Y, Li H, Wang X, Peng H, Yao Y, Hu Z, Ni Z, Xin M, Sun Q (2018b) Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat ( Triticum aestivum L.). Plant Biotechnol J 16:714–726
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco C, Register J, Zhao ZY, Xu D, Jones T, Gordon-Kamm W (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28:1998–2015
Lu Y, Loh Y-H, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, Collins JJ, Daley GQ, Marto JA (2014) Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell 15:92–101
Ma L, Liu M, Yan Y, Qing C, Zhang X, Zhang Y, Long Y, Wang L, Pan L, Zou C, Li Z, Wang Y, Peng H, Pan G, Jiang Z, Shen Y (2018) Genetic dissection of maize embryonic callus regenerative capacity using multi-locus genome-wide association studies. Front Plant Sci 9:561
Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22:1184–1195
Mei W, Liu S, Schnable JC, Yeh CT, Springer NM, Schnable PS, Barbazuk WB (2017) A comprehensive analysis of alternative splicing in paleopolyploid maize. Front Plant Sci 8:694
Ohta S, Nishida E, Yamanaka S, Yamamoto T (2013) Global splicing pattern reversion during somatic cell reprogramming. Cell Rep 5:357–366
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana : unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463
Opabode JT (2006) Agrobacterium-mediated transformation of plants: emerging factors that influence efficiency. Biotechnol Mol Biol Rev 1:12–20
Que Q, Elumalai S, Li X, Zhong H, Nalapalli S, Schweiner M, Fei X, Nuccio M, Kelliher T, Gu W, Chen Z, Chilton MD (2014) Maize transformation technology development for commercial event generation. Front Plant Sci 5:379
Reddy ASN, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25:3657–3683
Salvo SAGD, Hirsch CN, Buell CR, Kaeppler SM, Kaeppler HF (2014) Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS One 9:e111407
Sang YL, Cheng ZJ, Zhang XS (2018a) iPSCs: a comparison between animals and plants. Trends Plant Sci 23:660–666
Sang YL, Cheng ZJ, Zhang XS (2018b) Plant stem cells and de novo organogenesis. New Phytol 218:1334–1339
Shen Y, Jiang Z, Lu S, Lin H, Gao S, Peng H, Yuan G, Liu L, Zhang Z, Zhao M, Rong T, Pan G (2013) Combined small RNA and degradome sequencing reveals microRNA regulation during immature maize embryo dedifferentiation. Biochem Biophys Res Commun 441:425–430
Shen Y, Jiang Z, Yao X, Zhang Z, Lin H, Zhao M, Liu H, Peng H, Li S, Pan G (2012) Genome expression profile analysis of the immature maize embryo during dedifferentiation. PLoS One 7:e32237
Somssich M, Je BI, Simon R, Jackson D (2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143:3238–3248
Staiger D, Brown JWS (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25:3640–3656
Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18:463–471
Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat Commun 7:13274
Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW (2012) Alternative splicing in plants – coming of age. Trends Plant Sci 17:616–623
Thatcher SR, Danilevskaya ON, Meng X, Beatty M, Zastrow-Hayes G, Harris C, Van Allen B, Habben J, Li B (2016) Genome-wide analysis of alternative aplicing during development and drought stress in maize. Plant Physiol 170:586–599
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nucleic Acids Res 7:562–578
Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, Abou Elela S, Roux P, Lemaitre JM, Tazi J (2013) MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun 4:2480
von Arnold S, Sabala I, Bozhkov P, Dyachok J, Filonova L (2002) Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult 69:233–249
Wei Z-M, Huang X-Q (2004) High-frequency plant regeneration through callus initiation from mature embryos of maize (Zea Mays L.). Plant Cell Rep 22:793–800
Xu C, Cao H, Zhang Q, Wang H, Xin W, Xu E, Zhang S, Yu R, Yu D, Hu Y (2018) Control of auxin-induced callus formation by bZIP59–LBD complex in Arabidopsis regeneration. Nat Plants 4:108–115
Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25:2025–2030
Yadava P, Abhishek A, Singh R, Singh I, Kaul T, Pattanayak A, Agrawal PK (2017) Advances in maize transformation technologies and development of transgenic maize. Front Plant Sci 7:1949
Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC (2012) Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell 48:521–531
Zimmerman JL (1993) Somatic embryogenesis: a model for early development in higher plants. Plant Cell 5:1411–1423
Zouine M, Fu Y, Chateigner-Boutin AL, Mila I, Frasse P, Wang H, Audran C, Roustan JP, Bouzayen M (2014) Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS One 9:e84203
Acknowledgments
We are grateful to the reviewers for the valuable advice to improve the manuscript.
Funding
This work was supported by the National Major Project for Transgenic Organism Breeding (2016ZX08010-004), the Agricultural Science and Technology Innovation Program of CAAS and Youth Talent Plan of CAAS.
Author information
Authors and Affiliations
Contributions
XmD, TF, GyW, JjF, and YjL designed the research. XmD and TF performed the research. YL, LyH, YbC, and MsZ conducted the validation and collected the samples. TF, JZ, and XlW analyzed the data. XmD, TF, JjF, and YjL wrote the article.
Corresponding authors
Additional information
Editor: Yong Eui Choi
Rights and permissions
About this article
Cite this article
Du, X., Fang, T., Liu, Y. et al. Global Profiling of Alternative Splicing in Callus Induction of Immature Maize Embryo. In Vitro Cell.Dev.Biol.-Plant 56, 159–168 (2020). https://doi.org/10.1007/s11627-019-10024-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-019-10024-4