Abstract
Background
A high rate of postoperative recurrence, especially early recurrence (ER) occurring within 1 year, seriously impedes patients with hepatocellular carcinoma (HCC) from achieving long-term survival. This study aimed to establish a genomic-clinicopathologic nomogram for precisely predicting ER in HCC patients after R0 resection.
Methods
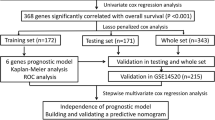
Two reliable datasets from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases were selected as the training and validation cohorts, respectively. The prognostic genes related to ER were screened out by univariate Cox regression analysis and differential expression analysis. The gene-based prognostic index was constructed using LASSO and Cox regression analyses, and its independent prognostic value was assessed by Kaplan-Meier and multivariate Cox analyses. Gene set enrichment analysis (GSEA) was performed to explore the biological pathways related to the prognostic index. Finally, the nomogram integrating all the independent prognostic factors was established and comprehensively evaluated by calibration plots, the C-index, receiver operating characteristic curves, and decision curve analysis.
Results
Nine dysregulated and prognostic genes related to ER (ZNF131, TATDN2, TXN, DDX55, KPNA2, ZNF30, TIMELESS, SFRP1, and COLEC11) were identified (all P < 0.05). The prognostic index model based on the 9 genes was successfully constructed using the TCGA cohort and showed a certain capability to discriminate the ER group from the non-ER group (P < 0.05) and good independent prognostic value in terms of predicting poor early recurrence-free survival (P < 0.05). Eight biological pathways significantly related to ER were identified by GSEA, such as “cell cycle”, “homologous recombination” and “p53 signaling pathway.” The genomic-clinicopathologic nomogram integrating the 9-gene-based prognostic index and TNM stage displayed significantly higher predictive accuracy and clinical application value than that of TNM stage model both in the training and validation cohorts (all P < 0.05).
Conclusions
The novel genomic-clinicopathologic nomogram may be a convenient and powerful tool for accurately predicting ER in HCC patients after R0 resection.







Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018; 391:1301–1314.
Akateh C, Black SM, Conteh L, Miller ED, Noonan A, Elliott E, Pawlik TM, Tsung A, Cloyd JM. Neoadjuvant and adjuvant treatment strategies for hepatocellular carcinoma. World J Gastroenterol 2019; 25:3704–3721.
Chan AWH, Zhong J, Berhane S, Toyoda H, Cucchetti A, Shi K, Tada T, Chong CCN, Xiang BD, Li LQ, Lai PBS, Mazzaferro V, Garcia-Finana M, Kudo M, Kumada T, Roayaie S, Johnson PJ. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol 2018; 69:1284–1293.
Cai J, Tong Y, Huang L, Xia L, Guo H, Wu H, Kong X, Xia Q. Identification and validation of a potent multi-mRNA signature for the prediction of early relapse in hepatocellular carcinoma. Carcinogenesis 2019; 40:840–852.
Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019; 156:477–491.e1.
Zhang XP, Chen ZH, Zhou TF, Li LQ, Chen MS, Wen TF, Shi J, Guo WX, Wu MC, Lau WY, Cheng SQ, Chinese National Research Cooperative Group for D, Treatment of Hepatocellular Carcinoma with Tumour T. A nomogram to predict early postoperative recurrence of hepatocellular carcinoma with portal vein tumour thrombus after R0 liver resection: A large-scale, multicenter study. Eur J Surg Oncol 2019; 45:1644–1651.
Zhang YM, Zhou ZT, Liu GM. Factors predicting early recurrence after surgical resection of hepatocellular carcinoma. J Hepatol 2019; 70:571–572.
Llovet JM, Di Bisceglie AM, Bruix J, Kramer BS, Lencioni R, Zhu AX, Sherman M, Schwartz M, Lotze M, Talwalkar J, Gores GJ, Panel of Experts in HCCDCT. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst 2008; 100:698–711.
Chan AC, Fan ST, Poon RT, Cheung TT, Chok KS, Chan SC, Lo CM. Evaluation of the seventh edition of the American Joint Committee on Cancer tumour-node-metastasis (TNM) staging system for patients undergoing curative resection of hepatocellular carcinoma: implications for the development of a refined staging system. HPB (Oxford) 2013; 15:439–448.
Addissie BD, Roberts LR. Classification and staging of hepatocellular carcinoma: an aid to clinical decision-making. Clin Liver Dis 2015; 19:277–294.
Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol 2019; 71:616–630.
Nault JC, Galle PR, Marquardt JU. The role of molecular enrichment on future therapies in hepatocellular carcinoma. J Hepatol 2018; 69:237–247.
Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnstrom H, Glimelius B, Sjoblom T, Edqvist PH, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science 2017; 357.
Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004; 10:7252–7259.
Wang Y, Tan PY, Handoko YA, Sekar K, Shi M, Xie C, Jiang XD, Dong QZ, Goh BKP, Ooi LL, Gao Z, Hui KM. NUF2 is a valuable prognostic biomarker to predict early recurrence of hepatocellular carcinoma after surgical resection. Int J Cancer 2019; 145:662–670.
Chen J, Xia H, Zhang X, Karthik S, Pratap SV, Ooi LL, Hong W, Hui KM. ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol 2015; 62:1287–1295.
Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL, Hong W, Hui KM. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/beta-catenin signalling pathway. Gut 2016; 65:1522–1534.
Xia H, Chen J, Shi M, Gao H, Sekar K, Seshachalam VP, Ooi LL, Hui KM. EDIL3 is a novel regulator of epithelial-mesenchymal transition controlling early recurrence of hepatocellular carcinoma. J Hepatol 2015; 63:863–873.
Donadon M, Di Tommaso L, Soldani C, Franceschini B, Terrone A, Mimmo A, Vitali E, Roncalli M, Lania A, Torzilli G. Filamin A expression predicts early recurrence of hepatocellular carcinoma after hepatectomy. Liver Int 2018; 38:303–311.
Wang YY, Qi LN, Zhong JH, Qin HG, Ye JZ, Lu SD, Ma L, Xiang BD, Li LQ, You XM. High expression of AKR1B10 predicts low risk of early tumor recurrence in patients with hepatitis B virus-related hepatocellular carcinoma. Sci Rep 2017; 7:42199.
Hayashi T, Ohtsuka M, Okamura D, Seki N, Kimura F, Shimizu H, Yoshidome H, Kato A, Yoshitomi H, Furukawa K, Miyazaki M. Cytoskeleton-associated protein 2 is a potential predictive marker for risk of early and extensive recurrence of hepatocellular carcinoma after operative resection. Surgery 2014; 155:114–123.
Yuan RH, Jeng YM, Pan HW, Hu FC, Lai PL, Lee PH, Hsu HC. Overexpression of KIAA0101 predicts high stage, early tumor recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2007; 13:5368–5376.
Kurokawa Y, Matoba R, Takemasa I, Nagano H, Dono K, Nakamori S, Umeshita K, Sakon M, Ueno N, Oba S, Ishii S, Kato K, Monden M. Molecular-based prediction of early recurrence in hepatocellular carcinoma. J Hepatol 2004; 41:284–291.
Yoshioka S, Takemasa I, Nagano H, Kittaka N, Noda T, Wada H, Kobayashi S, Marubashi S, Takeda Y, Umeshita K, Dono K, Matsubara K, Monden M. Molecular prediction of early recurrence after resection of hepatocellular carcinoma. Eur J Cancer 2009; 45:881–889.
Yuan S, Wang J, Yang Y, Zhang J, Liu H, Xiao J, Xu Q, Huang X, Xiang B, Zhu S, Li L, Liu J, Liu L, Zhou W. The Prediction of Clinical Outcome in Hepatocellular Carcinoma Based on a Six-Gene Metastasis Signature. Clin Cancer Res 2017; 23:289–297.
Zhang XY, Ou J, Chen JY, Li WW. Predicting early hepatocellular carcinoma recurrence after resection: A comment for moving forward. J Hepatol 2019; 70:567–568.
European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol 2018; 69:182–236.
Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018; 67:358–380.
Shim JH, Jun MJ, Han S, Lee YJ, Lee SG, Kim KM, Lim YS, Lee HC. Prognostic nomograms for prediction of recurrence and survival after curative liver resection for hepatocellular carcinoma. Ann Surg 2015; 261:939–946.
Ding Y, Herman JA, Toledo CM, Lang JM, Corrin P, Girard EJ, Basom R, Delrow JJ, Olson JM, Paddison PJ. ZNF131 suppresses centrosome fragmentation in glioblastoma stem-like cells through regulation of HAUS5. Oncotarget 2017; 8:48545–48562.
Jiang Y, Mei W, Gu Y, Lin X, He L, Zeng H, Wei F, Wan X, Yang H, Major P, Tang D. Construction of a set of novel and robust gene expression signatures predicting prostate cancer recurrence. Mol Oncol 2018; 12:1559–1578.
Sarkar M, Ghosh MK. DEAD box RNA helicases: crucial regulators of gene expression and oncogenesis. Front Biosci (Landmark Ed) 2016; 21:225–250.
Liu J, Wang D, Zhang C, Zhang Z, Chen X, Lian J, Liu J, Wang G, Yuan W, Sun Z, Wang W, Song M, Wang Y, Wu Q, Cao L, Wang D, Zhang Y. Identification of liver metastasis-associated genes in human colon carcinoma by mRNA profiling. Chin J Cancer Res 2018; 30:633–646.
Jiang M, Li M, Fu X, Huang Y, Qian H, Sun R, Mao Y, Xie Y, Li Y. Simultaneously detection of genomic and expression alterations in prostate cancer using cDNA microarray. Prostate 2008; 68:1496–1509.
Zhang C, Sun Q. Weighted gene co-expression network analysis of gene modules for the prognosis of esophageal cancer. J Huazhong Univ Sci Technolog Med Sci 2017; 37:319–325.
Bignotti E, Tassi RA, Calza S, Ravaggi A, Bandiera E, Rossi E, Donzelli C, Pasinetti B, Pecorelli S, Santin AD. Gene expression profile of ovarian serous papillary carcinomas: identification of metastasis-associated genes. Am J Obstet Gynecol 2007; 196:245 e1–11.
Cho SY, Kim S, Son MJ, Rou WS, Kim SH, Eun HS, Lee BS. Clinical Significance of the Thioredoxin System and Thioredoxin-Domain-Containing Protein Family in Hepatocellular Carcinoma. Dig Dis Sci 2019; 64:123–136.
Zhang T, Liu H, Zhu C, Briggs K, Kang Y, Fleming JA, Curley SA. Silencing thioredoxin induces liver cancer cell senescence under hypoxia. Hepatol Res 2012; 42:706–713.
Guo X, Wang Z, Zhang J, Xu Q, Hou G, Yang Y, Dong C, Liu G, Liang C, Liu L, Zhou W, Liu H. Upregulated KPNA2 promotes hepatocellular carcinoma progression and indicates prognostic significance across human cancer types. Acta Biochim Biophys Sin (Shanghai) 2019; 51:285–292.
Jiang P, Tang Y, He L, Tang H, Liang M, Mai C, Hu L, Hong J. Aberrant expression of nuclear KPNA2 is correlated with early recurrence and poor prognosis in patients with small hepatocellular carcinoma after hepatectomy. Med Oncol 2014; 31:131.
Gao CL, Wang GW, Yang GQ, Yang H, Zhuang L. Karyopherin subunit-alpha 2 expression accelerates cell cycle progression by upregulating CCNB2 and CDK1 in hepatocellular carcinoma. Oncol Lett 2018; 15:2815–2820.
Elgohary N, Pellegrino R, Neumann O, Elzawahry HM, Saber MM, Zeeneldin AA, Geffers R, Ehemann V, Schemmer P, Schirmacher P, Longerich T. Protumorigenic role of Timeless in hepatocellular carcinoma. Int J Oncol 2015; 46:597–606.
Wu Y, Li J, Sun CY, Zhou Y, Zhao YF, Zhang SJ. Epigenetic inactivation of the canonical Wnt antagonist secreted frizzled-related protein 1 in hepatocellular carcinoma cells. Neoplasma 2012; 59:326–332.
Davaadorj M, Imura S, Saito YU, Morine Y, Ikemoto T, Yamada S, Takasu C, Hiroki T, Yoshikawa M, Shimada M. Loss of SFRP1 Expression Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Anticancer Res 2016; 36:659–664.
Hu J, Dong A, Fernandez-Ruiz V, Shan J, Kawa M, Martinez-Anso E, Prieto J, Qian C. Blockade of Wnt signaling inhibits angiogenesis and tumor growth in hepatocellular carcinoma. Cancer Res 2009; 69:6951–6959.
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 81570079).
Author information
Authors and Affiliations
Contributions
Bin Yu designed this study, analyzed the data, and wrote the paper. Han Liang downloaded the data and prepared the data. Qifa Ye revised the paper. Yanfeng Wang designed this study, revised the paper, and provided supervision.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
Representative IHC images of the nine genes in the HPA database. (a) Protein levels of ZNF131 in normal liver tissues (https://www.proteinatlas.org/ENSG00000172262-ZNF131/tissue/liver#img) and HCC tissues (https://www.proteinatlas.org/ENSG00000172262-ZNF131/ pathology/liver+cancer#img). (b) Protein levels of TATDN2 in normal liver tissues (https://www.proteinatlas.org/ENSG00000157014-TATDN2/tissue/ liver#img) and HCC tissues (https://www.proteinatlas.org/ENSG00000157014- TATDN2/pathology/liver+cancer#img). (c) Protein levels of TXN in normal liver tissues (https://www.proteinatlas.org/ENSG00000136810-TXN/tissue/liver# img) and HCC tissues (https://www.proteinatlas.org/ENSG00000136810- TXN/pathology/liver+cancer#img). (d) Protein levels of DDX55 in normal liver tissues (https://www.proteinatlas.org/ENSG00000111364-DDX55/tissue/liver# img) and HCC tissues (https://www.proteinatlas.org/ENSG00000111364- DDX55/pathology/liver+cancer#img). (e) Protein levels of KPNA2 in normal liver tissues (https://www.proteinatlas.org/ENSG00000182481-KPNA2/tissue/ liver#img) and HCC tissues (https://www.proteinatlas.org/ENSG00000182481- KPNA2/pathology/liver+cancer#img). (f) Protein levels of ZNF30 in normal liver tissues (https://www.proteinatlas.org/ENSG00000168661-ZNF30/ tissue/liver#img) and HCC tissues (https://www.proteinatlas.org/ ENSG00000168661-ZNF30/pathology/liver+cancer#img). (g) Protein levels of TIMELESS in normal liver tissues (https://www.proteinatlas.org/ ENSG00000111602-TIMELESS/tissue/liver#img) and HCC tissues (https://www.proteinatlas.org/ENSG00000111602-TIMELESS/pathology/liver+cancer#img). (h) Protein levels of SFRP1 in normal liver tissues (https://www.proteinatlas.org/ENSG00000104332-SFRP1/tissue/liver#img) and HCC tissues (https://www.proteinatlas.org/ENSG00000104332-SFRP1/ pathology/liver+cancer#img). (i) Protein levels of COLEC11 in normal liver tissues (https://www.proteinatlas.org/ENSG00000118004-COLEC11/tissue /liver#img) and HCC tissues (https://www.proteinatlas.org/ ENSG00000118004-COLEC11/pathology/liver+cancer#img). In the HPA database, the expression level score is based on the staining intensity (negative, weak, moderate or strong) and fraction of stained cells (<25%, 25–75% or >75%), including not detected (N) (negative or weak with <25%), low (L) (weak with 25–75%/>75%, or moderate with <25%), medium (M) (moderate with 25–75%/>75%, or strong with <25%), and high (H) (strong with 25–75%/>75%). (PNG 2618 kb).
Supplementary Figure 2
Gene set enrichment analysis using the TCGA-LIHC dataset. A total of 8 KEGG signaling pathways were significantly enriched in high-risk group defined by gene-based prognostic index, including “cell cycle” (a), “DNA replication” (b), “base excision repair” (c), “spliceosome” (d), “nucleotide excision repair” (e), “RNA degradation” (f), “homologous recombination” (g) and “p53 signaling pathway” (h). NES: normalized enrichment score, NOM p: normalized p-value, FDR q: false discovery rate q-value. (PNG 1043 kb).
ESM 1
(DOCX 27.6 kb).
Rights and permissions
About this article
Cite this article
Yu, B., Liang, H., Ye, Q. et al. Establishment of a Genomic-Clinicopathologic Nomogram for Predicting Early Recurrence of Hepatocellular Carcinoma After R0 Resection. J Gastrointest Surg 25, 112–124 (2021). https://doi.org/10.1007/s11605-020-04554-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11605-020-04554-1