Abstract
Purpose
Ki67 is a protein associated with tumor proliferation and metastasis in breast cancer and acts as an essential prognostic factor. Clinical work requires recognizing tumor regions on Ki67-stained whole-slide images (WSIs) before quantitation. Deep learning has the potential to provide assistance but largely relies on massive annotations and consumes a huge amount of time and energy. Hence, a novel tumor region recognition approach is proposed for more precise Ki67 quantification.
Methods
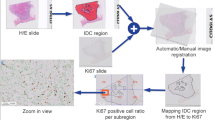
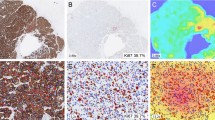
An unsupervised domain adaptive method is proposed, which combines adversarial and self-training. The model trained on labeled hematoxylin and eosin (H&E) data and unlabeled Ki67 data can recognize tumor regions in Ki67 WSIs. Based on the UDA method, a Ki67 automated assisted quantification system is developed, which contains foreground segmentation, tumor region recognition, cell counting, and WSI-level score calculation.
Results
The proposed UDA method achieves high performance in tumor region recognition and Ki67 quantification. The AUC reached 0.9915, 0.9352, and 0.9689 on the validation set and internal and external test sets, respectively, substantially exceeding baseline (0.9334, 0.9167, 0.9408) and rivaling the fully supervised method (0.9950, 0.9284, 0.9652). The evaluation of automated quantification on 148 WSIs illustrated statistical agreement with pathological reports.
Conclusion
The model trained by the proposed method is capable of accurately recognizing Ki67 tumor regions. The proposed UDA method can be readily extended to other types of immunohistochemical staining images. The results of automated assisted quantification are accurate and interpretable to provide assistance to both junior and senior pathologists in their interpretation.












Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Nielsen TO, Leung SCY, Rimm DL, Dodson A, Acs B, Badve S, Denkert C, Ellis MJ, Fineberg S, Flowers M, Kreipe HH, Laenkholm A-V, Pan H, Penault-Llorca FM, Polley M-Y, Salgado R, Smith IE, Sugie T, Bartlett JMS, McShane LM, Dowsett M, Hayes DF (2021) Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in breast cancer working group. JNCI 113(7):808–819. https://doi.org/10.1093/jnci/djaa201
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, https://arxiv.org/abs/1409.1556
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, 25. Commun. ACM 60, 6 June 2017, pp 84–90. https://doi.org/10.1145/3065386.
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778. https://ui.adsabs.harvard.edu/link_gateway/2015arXiv151203385H/arxiv:1512.03385.
Campanella G, Hanna MG, Geneslaw L, Miraflor A, Silva WK, Busam V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ (2019) Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 25(8):1301–1309. https://doi.org/10.1038/s41591-019-0508-1
Chen C-L, Chen C-C, Wei-Hsiang Yu, Chen S-H, Chang Y-C, Hsu T-I, Hsiao M, Yeh C-Y, Chen C-Y (2021) An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat Commun 12(1):1–13. https://doi.org/10.1038/s41467-021-21467-y
Lu MY, Chen TY, Williamson DF, Zhao M, Shady M, Lipkova J, Mahmood F (2021) AI-based pathology predicts origins for cancers of unknown primary. Nature 594(7861):106–110. https://doi.org/10.1038/s41586-021-03512-4
Choschzick M, Alyahiaoui M, Ciritsis A, Rossi C, Gut A, Hejduk P, Boss A (2021) Deep learning for the standardized classification of Ki-67 in vulva carcinoma: a feasibility study. Heliyon 7(7):e07577. https://doi.org/10.1016/j.heliyon.2021.e07577
Geread RS, Sivanandarajah A, Brouwer ER, Wood GA, Androutsos D, Faragalla H, Khademi A (2020) Pinet-an automated proliferation index calculator framework for Ki67 breast cancer images. Cancers 13(1):11. https://doi.org/10.3390/cancers13010011
Xing F, Cornish TC, Bennett T, Ghosh D, Yang L (2019) Pixel-to-pixel learning with weak supervision for single-stage nucleus recognition in Ki67 images. IEEE Trans Biomed Eng 66(11):3088–3097. https://doi.org/10.1109/TBME.2019.2900378
Razavi S, Khameneh FD, Serteli EA, Cayir S, Cetin SB, Hatipoglu G, Ayalti S, Kamasak M (2018) An automated and accurate methodology to assess ki-67 labeling index of immunohistochemical staining images of breast cancer tissues. In: 2018 25th international conference on systems, signals and image processing (IWSSIP). IEEE, 2018 June, pp 1–5. https://doi.org/10.1109/IWSSIP.2018.8439184.
Joseph J, Roudier MP, Narayanan PL, Augulis R, Ros VR, Pritchard A, Gerrard J, Laurinavicius A, Harrington EA, Carl Barrett J, Howat WJ (2019) Proliferation Tumour Marker Network (PTM-NET) for the identification of tumour region in Ki67 stained breast cancer whole slide images. Sci Rep 9(1):1–12. https://doi.org/10.1038/s41598-019-49139-4
Valkonen M, Isola J, Ylinen O, Muhonen V, Saxlin A, Tolonen T, Nykter M, Ruusuvuori P (2019) Cytokeratin-supervised deep learning for automatic recognition of epithelial cells in breast cancers stained for ER, PR, and Ki-67. IEEE Trans Med Imaging 39(2):534–542. https://doi.org/10.1109/TMI.2019.2933656
Valkonen M, Isola J, Ylinen O, Muhonen V, Saxlin A, Tolonen T, Nykter M, Ruusuvuori P (2022) Automated assessment of Ki-67 proliferation index in neuroendocrine tumors by deep learning. APMIS 130(1):11–20. https://doi.org/10.1111/apm.13190
Barricelli BR, Casiraghi E, Gliozzo J, Huber V, Leone BE, Rizzi A, Vergani B (2019) ki67 nuclei detection and ki67-index estimation: a novel automatic approach based on human vision modeling. BMC Bioinformatics 20(1):1–14. https://doi.org/10.1186/s12859-019-3285-4
Negahbani F, Sabzi R, Pakniyat Jahromi B, Firouzabadi D, Movahedi F, Shirazi K, Shirazi MK, Majidi S, Amirreza Dehghanian A (2021) PathoNet introduced as a deep neural network backend for evaluation of Ki-67 and tumor-infiltrating lymphocytes in breast cancer. Sci Rep 11(1):1–13. https://doi.org/10.1038/s41598-021-86912-w
Saha M, Chakraborty C, Arun I, Ahmed R, Chatterjee S (2017) An advanced deep learning approach for Ki-67 stained hotspot detection and proliferation rate scoring for prognostic evaluation of breast cancer. Sci Rep 7(1):1–14. https://doi.org/10.1038/s41598-017-03405-5
Feng M, Deng Y, Yang L, Jing Q, Zhang Z, Lian Xu, Wei X, Zhou Y, Diwei Wu, Xiang F, Wang Y, Bao Ji, Hong Bu (2020) Automated quantitative analysis of Ki-67 staining and HE images recognition and registration based on whole tissue sections in breast carcinoma. Diagn Pathol 15(1):1–12. https://doi.org/10.1186/s13000-020-00957-5
Weitz P, Acs B, Hartman J, Rantalainen M (2021) Prediction of Ki67 scores from H&E stained breast cancer sections using convolutional neural networks. https://openreview.net/pdf?id=W9sz0zHk33h
Niazi MKK, Senaras C, Pennell M, Arole V, Tozbikian G, Gurcan MN (2018) Relationship between the Ki67 index and its area based approximation in breast cancer. BMC Cancer 18(1):1–9. https://doi.org/10.1186/s12885-018-4735-5
Ganin Y, Ustinova E, Ajakan H, Germain P, Larochelle H, Laviolette F, Marchand M, Lempitsky V (2016) Domain-adversarial training of neural networks. J Mach Learn Res 17(1):2096–2030. https://doi.org/10.1007/978-3-319-58347-1
Tzeng E, Hoffman J, Saenko K, Darrell T (2017) Adversarial discriminative domain adaptation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7167–7176. https://ui.adsabs.harvard.edu/link_gateway/2017arXiv170205464T/arXiv:1702.05464.
Scudder H (1965) Probability of error of some adaptive pattern-recognition machines. IEEE Trans Inf Theory 11(3):363–371. https://doi.org/10.1109/TIT.1965.1053799
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, 2015 Oct, pp 234–241 https://doi.org/10.1007/978-3-319-24574-4_28.
Zhang R, Yang J, Chen C (2018) Tumor cell identification in ki-67 images on deep learning. Mol Cell Biomech 15(3):177. https://doi.org/10.3970/mcb.2018.04292
Glenn Jocher, Alex Stoken, Jirka Borovec, NanoCode012, ChristopherSTAN, Liu Changyu, Laughing, Adam Hogan, lorenzomammana, tkianai, yxNONG, AlexWang1900, Laurentiu Diaconu, Marc, wanghaoyang0106, ml5ah, Doug, Hatovix, Jake Poznanski, Lijun Yu, changyu98, Prashant Rai, Russ Ferriday, Trevor Sullivan, Wang Xinyu, YuriRibeiro, Eduard Reñé Claramunt, hopesala, pritul dave & yzchen (2020) ultralytics/yolov5: v3.0 (v3.0). Zenodo. https://zenodo.org/record/3983579/export/geojson#.Y232THZByUk
Paik S, Kwon Y, Lee MH, Kim JY, Lee DK, Cho WJ, Lee EY, Lee ES (2021) Systematic evaluation of scoring methods for Ki67 as a surrogate for 21-gene recurrence score. NPJ Breast Cancer 7(1):1–8
Robertson S, Acs B, Lippert M, Hartman J (2020) Prognostic potential of automated Ki67 evaluation in breast cancer: different hot spot definitions versus true global score. Breast Cancer Res Treat 183(1):161–175. https://doi.org/10.1007/s10549-020-05752-w
Reis-Filho JS, Davidson NE (2021) Ki67 assessment in breast cancer: are we there yet? JNCI 113(7):797–798. https://doi.org/10.1093/jnci/djaa202
Acknowledgements
Authors acknowledge the pathologists and assistants participating in this study for specimen collection, preparation, and quality control at the Shenzhen Center, Cancer Hospital Chinese Academy of Medical Sciences and Huaibei Maternal and child Health Care Hospital. This work was supported by National Science Foundation of China (61875102), Science and Technology Research Program of Shenzhen City (JCYJ20200109110606054), Science and Technology Research Program of Shenzhen City (JCYJ20180508152528735), Sanming Project of Medicine in Shenzhen(No.SZSM201812076), Shenzhen High-level Hosiptal Construction Fund and Tsinghua University Spring Breeze Fund (2020Z99CFZ023). None declared conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. For this type of study, formal consent is not required. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, Q., Liu, Y., Pan, F. et al. Unsupervised domain adaptive tumor region recognition for Ki67 automated assisted quantification. Int J CARS 18, 629–640 (2023). https://doi.org/10.1007/s11548-022-02781-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02781-2