Abstract
Purpose
We address the automatic segmentation of healthy and cancerous liver tissues (parenchyma, active and necrotic parts of hepatocellular carcinoma (HCC) tumor) on multiphase CT images using a deep learning approach.
Methods
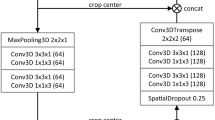
We devise a cascaded convolutional neural network based on the U-Net architecture. Two strategies for dealing with multiphase information are compared: Single-phase images are concatenated in a multi-dimensional features map on the input layer, or output maps are computed independently for each phase before being merged to produce the final segmentation. Each network of the cascade is specialized in the segmentation of a specific tissue. The performances of these networks taken separately and of the cascaded architecture are assessed on both single-phase and on multiphase images.
Results
In terms of Dice coefficients, the proposed method is on par with a state-of-the-art method designed for automatic MR image segmentation and outperforms previously used technique for interactive CT image segmentation. We validate the hypothesis that several cascaded specialized networks have a higher prediction accuracy than a single network addressing all tasks simultaneously. Although the portal venous phase alone seems to provide sufficient contrast for discriminating tumors from healthy parenchyma, the multiphase information brings significant improvement for the segmentation of cancerous tissues (active versus necrotic part).
Conclusion
The proposed cascaded multiphase architecture showed promising performances for the automatic segmentation of liver tissues, allowing to reliably estimate the necrosis rate, a valuable imaging biomarker of the clinical outcome.







Similar content being viewed by others
References
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Ben-Cohen A, Diamant I, Klang E, Amitai M, Greenspan H (2016) Fully convolutional network for liver segmentation and lesions detection. In: Carneiro G et al (eds) Deep learning and data labeling for medical applications, vol 10008. Springer, Cham, pp 77–85. https://doi.org/10.1007/978-3-319-46976-8
Christ PF, Ettlinger F, Grün F, Elshaera MEA, Lipkova J, Schlecht S, Ahmaddy F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Hofmann F, Anastasi MD, Ahmadi SA, Kaissis G, Holch J, Sommer W, Braren R, Heinemann V, Menze B (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks, pp 1–20. arXiv:1702.05970
Conze PH, Noblet V, Rousseau F, Heitz F, de Blasi V, Memeo R, Pessaux P (2017) Scale-adaptive supervoxel-based random forests for liver tumor segmentation in dynamic contrast-enhanced CT scans. Int J Comput Assist Radiol Surg 12(2):223–233. https://doi.org/10.1007/s11548-016-1493-1
Erdt M, Steger S, Kirschner M, Wesarg S (2010) Fast automatic liver segmentation combining learned shape priors with observed shape deviation. In: Proceedings—IEEE symposium on computer-based medical systems, pp 249–254. https://doi.org/10.1109/CBMS.2010.6042650
Bray F, Ferlay Jacques, Soerjomataram Isabelle, Siegel RL, Torre LA, Jemal A (2018) Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. https://doi.org/10.3322/caac.21492
Foruzan AH, Aghaeizadeh Zoroofi R, Hori M, Sato Y (2009) Liver segmentation by intensity analysis and anatomical information in multi-slice CT images. Int J Comput Assist Radiol Surg 4(3):287–297. https://doi.org/10.1007/s11548-009-0293-2
Freiman M, Cooper O, Lischinski D, Joskowicz L (2011) Liver tumors segmentation from CTA images using voxels classification and affinity constraint propagation. Int J Comput Assist Radiol Surg 6(2):247–255. https://doi.org/10.1007/s11548-010-0497-5
Hu Z, Tang J, Wang Z, Zhang K, Zhang L, Sun Q (2018) Deep learning for image-based cancer detection and diagnosis : a survey. Pattern Recognit 83:134–149. https://doi.org/10.1016/j.patcog.2018.05.014
Jeong WK, Jamshidi N, Felker ER, Raman SS, Lu DS (2018) Radiomics and radiogenomics of primary liver cancers. Clin Mol Hepatol 25:1–9. https://doi.org/10.3350/cmh.2018.1007
Ker J, Wang L, Rao J, Lim T (2017) Deep learning applications in medical image analysis. IEEE Access 6:9375–9379. https://doi.org/10.1109/ACCESS.2017.2788044
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. In: AIP conference proceedings 1631:58–62. https://doi.org/10.1063/1.4902458. arXiv:1412.6980
Kumar SS, Moni RS, Rajeesh J (2013) Automatic liver and lesion segmentation: a primary step in diagnosis of liver diseases. Signal Image Video Process 7(1):163–172. https://doi.org/10.1007/s11760-011-0223-y
Lee J, Kim KW, Kim SY, Shin J, Park KJ, Won HJ, Shin YM (2015) Automatic detection method of hepatocellular carcinomas using the non-rigid registration method of multi-phase liver CT images. J X-ray Sci Technol 23(3):275–288. https://doi.org/10.3233/XST-150487
Li CY, Wang X, Eberl S, Fulham M, Yin Y, Feng D (2010) Fully automated liver segmentation for low- and high-contrast ct volumes based on probabilistic atlases. In: Proceedings—international conference on image processing ICIP, pp 1733–1736. https://doi.org/10.1109/ICIP.2010.5654434
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42(1995):60–88. https://doi.org/10.1016/j.media.2017.07.005
Moghbel M, Mashohor S, Mahmud R, Saripan MIB (2018) Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev 50(4):497–537. https://doi.org/10.1007/s10462-017-9550-x
Novikov AA, Lenis D, Major D, Hladuvka J, Wimmer M, Buhler K (2018) Fully convolutional architectures for multi-class segmentation in chest radiographs. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2018.2806086
O’Connor JPB, Rose CJ, Waterton JC, Carano RAD, Parker GJM, Jackson A (2015) Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res 21(2):249–257. https://doi.org/10.1158/1078-0432.CCR-14-0990
Oldhafer KJ, Chavan A, Frühauf NR, Flemming P, Schlitt HJ, Kubicka S, Nashan B, Weimann A, Raab R, Manns MP, Galanski M (1998) Arterial chemoembolization before liver transplantation in patients with hepatocellular carcinoma: Marked tumor necrosis, but no survival benefit? J Hepatol 29(6):953–959. https://doi.org/10.1016/S0168-8278(98)80123-2
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation, pp 1–8. arXiv:1505.04597
Ruskó L, Bekes G, Fidrich M (2009) Automatic segmentation of the liver from multi- and single-phase contrast-enhanced CT images. Med Image Anal 13(6):871–882. https://doi.org/10.1016/j.media.2009.07.009
Sadigh G, Applegate KE, Baumgarten DA (2014) Comparative accuracy of intravenous contrast-enhanced CT versus noncontrast CT plus intravenous contrast-enhanced CT in the detection and characterization of patients with hypervascular liver metastases. Critic Apprais Topic Acad Radiol 21(1):113–125. https://doi.org/10.1016/j.acra.2013.08.023
Saito A, Yamamoto S, Nawano S, Shimizu A (2017) Automated liver segmentation from a postmortem CT scan based on a statistical shape model. Int J Comput Assist Radiol Surg 12(2):205–221. https://doi.org/10.1007/s11548-016-1481-5
Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD (2007) Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 25(6):675–680. https://doi.org/10.1038/nbt1306
Shi C, Cheng Y, Liu F, Wang Y, Bai J, Tamura S (2016) A hierarchical local region-based sparse shape composition for liver segmentation in CT scans. Pattern Recognit 50:88–106. https://doi.org/10.1016/j.patcog.2015.09.001
Sun C, Guo S, Zhang H, Li J, Chen M, Ma S, Jin L, Liu X, Li X, Qian X (2017) Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs. Artif Intell Med 83:58–66. https://doi.org/10.1016/j.artmed.2017.03.008
Suzuki K (2017) Overview of deep learning in medical imaging. Radiol Phys Technol 10(3):257–273. https://doi.org/10.1007/s12194-017-0406-5
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23(7):903–921. https://doi.org/10.1109/TMI.2004.828354
Zhang F, Yang J, Nezami N, Laage-gaupp F, Chapiro J, De Lin M, Duncan J (2018) Liver tissue classification using an auto-context-based deep neural network with a multi-phase training framework. In: Bai W, Sanroma G, Wu G, Munsell BC, Zhan Y, Coupé P (eds) Patch-based techniques in medical imaging. Springer, Cham, pp 59–66
Zheng W, Thorne N, Mckew JC (2015) Deep learning in medical image analysis. Annu Rev Biomed Eng 18:1067–1073. https://doi.org/10.1016/j.drudis.2013.07.001.Phenotypic
Funding
This study was funded by IHU Strasbourg through ANR grant 10-IAHU-0002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ouhmich, F., Agnus, V., Noblet, V. et al. Liver tissue segmentation in multiphase CT scans using cascaded convolutional neural networks. Int J CARS 14, 1275–1284 (2019). https://doi.org/10.1007/s11548-019-01989-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-01989-z