Abstract
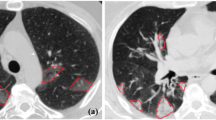
The ongoing COronaVIrus Disease 2019 (COVID-19) pandemic carried by the SARS-CoV-2 virus spread worldwide in early 2019, bringing about an existential health catastrophe. Automatic segmentation of infected lungs from COVID-19 X-ray and computer tomography (CT) images helps to generate a quantitative approach for treatment and diagnosis. The multi-class information about the infected lung is often obtained from the patient’s CT dataset. However, the main challenge is the extensive range of infected features and lack of contrast between infected and normal areas. To resolve these issues, a novel Global Infection Feature Network (GIFNet)-based Unet with ResNet50 model is proposed for segmenting the locations of COVID-19 lung infections. The Unet layers have been used to extract the features from input images and select the region of interest (ROI) by using the ResNet50 technique for training it faster. Moreover, integrating the pooling layer into the atrous spatial pyramid pooling (ASPP) mechanism in the bottleneck helps for better feature selection and handles scale variation during training. Furthermore, the partial differential equation (PDE) approach is used to enhance the image quality and intensity value for particular ROI boundary edges in the COVID-19 images. The proposed scheme has been validated on two datasets, namely the SARS-CoV-2 CT scan and COVIDx-19, for detecting infected lung segmentation (ILS). The experimental findings have been subjected to a comprehensive analysis using various evaluation metrics, including accuracy (ACC), area under curve (AUC), recall (REC), specificity (SPE), dice similarity coefficient (DSC), mean absolute error (MAE), precision (PRE), and mean squared error (MSE) to ensure rigorous validation. The results demonstrate the superior performance of the proposed system compared to the state-of-the-art (SOTA) segmentation models on both X-ray and CT datasets.
Graphical abstract
















Similar content being viewed by others
Data availability
The data that support the finding made in this manuscript can be accessed via the following link: https://github.com/HzFu/COVID19_imaging_AI_paper_list for SARS-CoV-2 CT scan dataset and link: https://www.kaggle.com/datasets/prashant268/chest-xray-COVID19-pneumonia for COVIDx-19 dataset.
References
Johns Hopkins University (2021) COVID-19 dashboard. [Online]. Available: https://coronavirus.jhu.edu/map.html Accessed Jun 10 2023
Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Xia L (2020) Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 296(2):32–40. https://doi.org/10.1148/radiol.2020200642
Lyu F, Ye M, Carlsen JF, Erleben K, Darkner S, Yuen PC (2022) Pseudo-label guided image synthesis for semi-supervised COVID-19 pneumonia infection segmentation. IEEE Trans Med Imaging 42(3):797–809. https://doi.org/10.1109/TMI.2022.3217501
Murmu A, Kumar P (2021) Deep learning model-based segmentation of medical diseases from MRI and CT images. In: TENCON 2021 IEEE Region 10 Conference (TENCON): pp 608–613. https://doi.org/10.1109/TENCON54134.2021.9707278
Sailunaz K, Özyer T, Rokne J, Alhajj R, (2023) A survey of machine learning-based methods for COVID-19 medical image analysis. Med Biol Eng Compu 1–41. https://doi.org/10.1007/s11517-022-02758-y
Yao Q, Xiao L, Liu P, Zhou SK (2021) Label-free segmentation of COVID-19 lesions in lung CT. IEEE Trans Med Imaging 40(10):2808–2819. https://doi.org/10.1109/TMI.2021.3066161
Murmu A, Kumar P (2023) A novel gateaux derivatives with efficient DCNN-ResUNet method for segmenting multi-class brain tumor. Med Biol Eng Compu. https://doi.org/10.1007/s11517-023-02824-z. (In press)
Zhang J, Chen D, Ma D, Ying C, Sun X, Xu X, Cheng Y (2023) CdcSegNet: automatic COVID-19 infection segmentation from CT images. IEEE Trans Instrum Meas 72. https://doi.org/10.1109/TIM.2023.3267355
Cobes N, Guernou M, Lussato D, Queneau M, Songy B, Bonardel G, Grellier JF (2020) Ventilation/perfusion SPECT/CT findings in different lung lesions associated with COVID-19: a case series. Eur J Nucl Med Mol Imaging 47(10):2453–2460. https://doi.org/10.1007/s00259-020-04920-w
Abdel-Basset M, Chang V, Mohamed R (2020) HSMA_WOA: a hybrid novel Slime mould algorithm with whale optimization algorithm for tackling the image segmentation problem of chest X-ray images. Appl Soft Comput 95:106642. https://doi.org/10.1016/j.asoc.2020.106642
Fan DP, Zhou T, Ji GP, Zhou Y, Chen G, Fu H, Shao L (2020) Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Med Imaging 39(8):2626–2637. https://doi.org/10.1109/TMI.2020.2996645
Qiu Y, Liu Y, Li S, Xu J (2021) MiniSeg: an extremely minimum network for efficient COVID-19 segmentation. Proceed AAAI Conf Artificial Intell 35(6):4846–4854. https://doi.org/10.1609/aaai.v35i6.16617
Yan Q, Wang B, Gong D, Luo C, Zhao W, Shen J, You Z (2020) COVID-19 chest CT image segmentation–a deep convolutional neural network solution. arXiv:2004.10987
Saeedizadeh N, Minaee S, Kafieh R, Yazdani S, Sonka M (2021) COVID TV-Unet: segmenting COVID-19 chest CT images using connectivity imposed Unet. Comput method Prog in Biomed Update 1:100007. https://doi.org/10.1016/j.cmpbup.2021.100007
Fan C, Zeng Z, Xiao L, Qu X (2022) GFNet: automatic segmentation of COVID-19 lung infection regions using CT images based on boundary features. Pattern Recogn 132:108963. https://doi.org/10.1016/j.patcog.2022.108963
Bougourzi F, Distante C, Dornaika F, Taleb-Ahmed A (2023) PDAtt-Unet: pyramid dual-decoder attention Unet for COVID-19 infection segmentation from CT-scans. Med Image Anal 86:102797. https://doi.org/10.1016/j.media.2023.102797
Roth HR, Xu Z, Tor-Díez C, Jacob RS, Zember J, Molto J, Linguraru MG (2022) Rapid artificial intelligence solutions in a pandemic-the COVID-19-20 lung CT lesion segmentation challenge. Med Image Anal 82:102605. https://doi.org/10.1016/j.media.2022.102605
Zhang Y, Liao Q, Yuan L, Zhu H, Xing J, Zhang J (2021) Exploiting shared knowledge from non-COVID lesions for annotation-efficient COVID-19 CT lung infection segmentation. IEEE J Biomed Health Inform 25(11):4152–4162. https://doi.org/10.1109/JBHI.2021.3106341
Wang G, Liu X, Li C, Xu Z, Ruan J, Zhu H, Zhang S (2020) A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans Med Imaging 39(8):2653–2663. https://doi.org/10.1109/TMI.2020.3000314
Liu J, Dong B, Wang S, Cui H, Fan DP, Ma J, Chen G (2021) COVID-19 lung infection segmentation with a novel two-stage cross-domain transfer learning framework. Med Image Anal 74:102205. https://doi.org/10.1016/j.media.2021.102205
Roy K, Banik D, Bhattacharjee D, Krejcar O, Kollmann C (2022) LwMLA-NET: a lightweight multi-level attention-based network for segmentation of COVID-19 lungs abnormalities from CT images. IEEE Trans Instrum Meas 71:1–13. https://doi.org/10.1109/TIM.2022.3161690
Chen H, Jiang Y, Ko H, Loew M (2023) A teacher-student framework with Fourier Transform augmentation for COVID-19 infection segmentation in CT images. Biomed Signal Process Control 79:104250. https://doi.org/10.1016/j.bspc.2022.104250
Rao Y, Lv Q, Zeng S, Yi Y, Huang C, Gao Y, Sun J (2023) COVID-19 CT ground-glass opacity segmentation based on attention mechanism threshold. Biomed Signal Process Control 81:104486. https://doi.org/10.1016/j.bspc.2022.104486
Khan A, Garner R, Rocca ML, Salehi S, Duncan D (2023) A novel threshold-based segmentation method for quantification of COVID-19 lung abnormalities. SIViP 17(4):907–914. https://doi.org/10.1007/s11760-022-02183-6
Fu H, Fan D-P, Chen G, Zhou T (2020) COVID-19 imaging-based AI research collection. Accessed 27 May 2020. [Online]. [Available online: https://github.com/HzFu/COVID19_imaging_AI_paper_list] [Accessed 15 Jun 2023
COVID-19 radiography database [Available online: https://www.kaggle.com/datasets/prashant268/chest-xray-COVID19-pneumonia] Accessed Jun 15 2023
Kumar P, Agrawal A (2015) Hardware accelerated multi-coordinate viewing framework for volumetric visualization of large 3D medical dataset. Procedia Comput Sci 54:566–573. https://doi.org/10.1016/j.procs.2015.06.065
Demirel H, Anbarjafari G (2011) Discrete wavelet transform-based satellite image resolution enhancement. IEEE Trans Geosci Remote Sens 49(6):1997–2004. https://doi.org/10.1109/TGRS.2010.2100401
Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV). pp 3-19
Saeedizadeh N, Minaee S, Kafieh R, Yazdani S, Sonka M (2021) COVID TV-Unet: segmenting COVID-19 chest CT images using connectivity imposed Unet. Comput Method Prog Biomed Update 1:100007. https://doi.org/10.1016/j.cmpbup.2021.100007
Cong R, Yang H, Jiang Q, Gao W, Li H, Wang C, Kwong S (2022) BCS-Net: boundary, context, and semantic for automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Instrum Meas 71:1–11. https://doi.org/10.1109/TIM.2022.3196430
Cong R, Zhang Y, Yang N, Li H, Zhang X, Li R, Kwong S (2022) Boundary guided semantic learning for real-time COVID-19 lung infection segmentation system. IEEE Trans Consum Electron 68(4):376–386. https://doi.org/10.1109/TCE.2022.3205376
Author information
Authors and Affiliations
Contributions
Anita Murmu (AnM) designed the environmental/implementation details platform, performed statistical analyses, and conducted experiments. Piyush Kumar (PiK) authored the literature survey and abstract section. AnM and PiK collaborated on writing the initial draft of the manuscript. PiK edited the first draft of the paper, while both authors contributed to analysis and result framing. Both authors reviewed and approved the final version of the manuscript. It is important to note that both Anita Murmu and Piyush Kumar made equal and significant contributions to this work.
Corresponding author
Ethics declarations
Ethical approval
This section is not relevant to this paper as it does not involve any studies with human participants conducted by the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Murmu, A., Kumar, P. GIFNet: an effective global infection feature network for automatic COVID-19 lung lesions segmentation. Med Biol Eng Comput (2024). https://doi.org/10.1007/s11517-024-03024-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11517-024-03024-z