Abstract
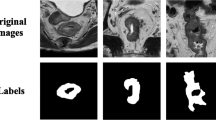
Accurate segmentation of rectal tumors is the most crucial task in determining the stage of rectal cancer and developing suitable therapies. However, complex image backgrounds, irregular edge, and poor contrast hinder the related research. This study presents an attention-based multi-modal fusion module to effectively integrate complementary information from different MRI images and suppress redundancy. In addition, a deep learning–based segmentation model (AF-UNet) is designed to achieve accurate segmentation of rectal tumors. This model takes multi-parametric MRI images as input and effectively integrates the features from different multi-parametric MRI images by embedding the attention fusion module. Finally, three types of MRI images (T2, ADC, DWI) of 250 patients with rectal cancer were collected, with the tumor regions delineated by two oncologists. The experimental results show that the proposed method is superior to the most advanced image segmentation method with a Dice coefficient of \(0.821\pm 0.065\), which is also better than other multi-modal fusion methods.
Graphical Abstract
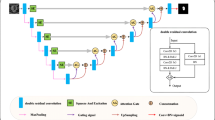
Framework of the AF-UNet. This model takes multi-modal MRI images as input, and integrates complementary information using attention mechanism and suppresses redundancy.









Similar content being viewed by others
References
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Feng L, Liu Z, Li C et al (2022) Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health 4:e8–e17. https://doi.org/10.1016/S2589-7500(21)00215-6
Bi WL, Hosny A, Schabath MB et al (2019) Artificial intelligence in cancer imaging: clinical challenges and applications. CA A Cancer J Clin caac.21552. https://doi.org/10.3322/caac.21552
Shin J, Seo N, Baek S-E et al (2022) MRI radiomics model predicts pathologic complete response of rectal cancer following chemoradiotherapy. Radiol 211986. https://doi.org/10.1148/radiol.211986
van Heeswijk (2017) Novel MR techniques in rectal cancer: translating research into practice. Maastricht University
Men K, Boimel P, Janopaul-Naylor J et al (2018) Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol 63:185016. https://doi.org/10.1088/1361-6560/aada6c
Zheng B, Cai C, Ma L (2020) CT images segmentation method of rectal tumor based on modified U-Net. 2020 16th International Conference on Control, Automation, Robotics and Vision (ICARCV). IEEE, Shenzhen, China, pp 672–677
Wang Y, Xu C (2021) CT image segmentation of rectal tumor based on U-Net network. 2021 2nd International Conference on Big Data and Informatization Education (ICBDIE). IEEE, Hangzhou, China, pp 69–72
Zhu H, Zhang X, Shi Y et al (2021) Automatic segmentation of rectal tumor on diffusion-weighted images by deep learning with U-Net. J Appl Clin Med Phys 22:324–331. https://doi.org/10.1002/acm2.13381
Rao Y, Zheng W, Zeng S, Sun J (2021) Using channel concatenation and lightweight atrous convolution in U-Net for accurate rectal cancer segmentation. 2021 4th International Conference on Pattern Recognition and Artificial Intelligence (PRAI). IEEE, Yibin, China, pp 247–254
Pei Y, Mu L, Fu Y et al (2020) Colorectal tumor segmentation of CT scans based on a convolutional neural network with an attention mechanism. IEEE Access 8:64131–64138. https://doi.org/10.1109/ACCESS.2020.2982543
Zhou T, Ruan S, Canu S (2019) A review: deep learning for medical image segmentation using multi-modality fusion. Array 3–4:100004. https://doi.org/10.1016/j.array.2019.100004
Zhang W, Li R, Deng H et al (2015) Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage 108:214–224. https://doi.org/10.1016/j.neuroimage.2014.12.061
Chen H, Dou Q, Yu L et al (2018) VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 170:446–455. https://doi.org/10.1016/j.neuroimage.2017.04.041
Nie D, Wang L, Gao Y, Shen D (2016) Fully convolutional networks for multi-modality isointense infant brain image segmentation. In 2016 IEEE 13Th international symposium on biomedical imaging (ISBI) (1342–1345). IEEE.
Li X, Dou Q, Chen H et al (2018) 3D multi-scale FCN with random modality voxel dropout learning for intervertebral disc localization and segmentation from multi-modality MR images. Med Image Anal 45:41–54. https://doi.org/10.1016/j.media.2018.01.004
Wang S, Burtt K, Turkbey B et al (2014) Computer aided-diagnosis of prostate cancer on multiparametric MRI: a technical review of current research. Biomed Res Int 2014:1–11. https://doi.org/10.1155/2014/789561
Dolz J, Gopinath K, Yuan J, Lombaert H, Desrosiers C, Ayed IB (2018) HyperDense-Net: a hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans Med Imaging 38(5):1116–1126
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds) Medical image computing and computer-assisted intervention – MICCAI 2015. Springer International Publishing, Cham, pp 234–241
Diakogiannis FI, Waldner F, Caccetta P, Wu C (2020) ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. ISPRS J Photogramm Remote Sens 162:94–114. https://doi.org/10.1016/j.isprsjprs.2020.01.013
Oktay O, Schlemper J, Folgoc LL et al (2018) Attention U-Net: learning where to look for the pancreas. arXiv:180403999 [cs]
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J (2018) UNet++: a nested U-Net architecture for medical image segmentation. In Deep learning in medical image analysis and multimodal learning for clinical decision support (3–11). Springer, Cham.
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H (2019) Dual attention network for scene segmentation. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (3146–3154).
Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV) (801–818).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) under Grant No. 82073338 and Sichuan Province Science and Technology Support Program No. 2022YFS0217.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committee of West China Hospital, Sichuan University, and all participants of the study completed an informed consent form prior to participation.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dou, M., Chen, Z., Tang, Y. et al. Segmentation of rectal tumor from multi-parametric MRI images using an attention-based fusion network. Med Biol Eng Comput 61, 2379–2389 (2023). https://doi.org/10.1007/s11517-023-02828-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02828-9