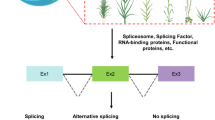
Abstract
Intron retention is the most common alternative splicing event in plants and plays a crucial role in the responses of plants to environmental signals. Despite a large number of RNA-seq libraries from different treatments and genetic mutants stored in public domains, a resource for querying the intron-splicing ratio of individual intron is still required. Here, we established the first-ever large-scale splicing efficiency database in any organism. Our database includes over 57,000 plant public RNA-seq libraries, comprising 25,283 from Arabidopsis, 17,789 from maize, 10,710 from rice, and 3,974 from soybean, and covers a total of 1.6 million introns in these four species. In addition, we manually curated and annotated all the mutant- and treatment-related libraries as well as their matched controls included in our library collection, and added graphics to display intron-splicing efficiency across various tissues, developmental stages, and stress-related conditions. The result is a large collection of 3,313 treatment conditions and 3,594 genetic mutants for discovering differentially regulated splicing efficiency. Our online database can be accessed at https://plantintron.com/.
Similar content being viewed by others
References
Chamala, S., Feng, G., Chavarro, C., and Barbazuk, W.B. (2015). Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front Bioeng Biotechnol 3, 33.
Chaudhary, S., Khokhar, W., Jabre, I., Reddy, A.S.N., Byrne, L.J., Wilson, C.M., and Syed, N.H. (2019). Alternative splicing and protein diversity: plants versus animals. Front Plant Sci 10, 708.
Cheng, C., Wang, Z., Yuan, B., and Li, X. (2017). RBM25 mediates abiotic responses in plants. Front Plant Sci 8.
Deng, X., Lu, T., Wang, L., Gu, L., Sun, J., Kong, X., Liu, C., and Cao, X. (2016). Recruitment of the NineTeen complex to the activated spliceosome requires AtPRMT5. Proc Natl Acad Sci USA 113, 5447–5452.
Dong, C., He, F., Berkowitz, O., Liu, J., Cao, P., Tang, M., Shi, H., Wang, W., Li, Q., Shen, Z., et al. (2018). Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 30, 2267–2285.
Filichkin, S.A., Hamilton, M., Dharmawardhana, P.D., Singh, S.K., Sullivan, C., Ben-Hur, A., Reddy, A.S.N., and Jaiswal, P. (2018). Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Front Plant Sci 9, 5.
Jacob, A.G., and Smith, C.W.J. (2017). Intron retention as a component of regulated gene expression programs. Hum Genet 136, 1043–1057.
Jia, J., Long, Y., Zhang, H., Li, Z., Liu, Z., Zhao, Y., Lu, D., Jin, X., Deng, X., Xia, R., et al. (2020). Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat Plants 6, 780–788.
Kim, D., Langmead, B., and Salzberg, S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360.
Kornblihtt, A.R., Schor, I.E., Alló, M., Dujardin, G., Petrillo, E., and Muñoz, M.J. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14, 153–165.
Laloum, T., Martín, G., and Duque, P. (2018). Alternative splicing control of abiotic stress responses. Trends Plant Sci 23, 140–150.
Lee, S.K., Eom, J.S., Hwang, S.K., Shin, D., An, G., Okita, T.W., and Jeon, J.S. (2016). Plastidic phosphoglucomutase and ADP-glucose pyrophosphorylase mutants impair starch synthesis in rice pollen grains and cause male sterility. J Exp Bot 67, 5557–5569.
Love, M.I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550.
Marquez, Y., Brown, J.W.S., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22, 1184–1195.
Martín, G., Márquez, Y., Mantica, F., Duque, P., and Irimia, M. (2021). Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol 22, 35.
Mei, W., Boatwright, L., Feng, G., Schnable, J.C., and Barbazuk, W.B. (2017a). Evolutionarily conserved alternative splicing across monocots. Genetics 207, 465–480.
Mei, W., Liu, S., Schnable, J.C., Yeh, C.T., Springer, N.M., Schnable, P.S., and Barbazuk, W.B. (2017b). A comprehensive analysis of alternative splicing in paleopolyploid maize. Front Plant Sci 8, 694.
Middleton, R., Gao, D., Thomas, A., Singh, B., Au, A., Wong, J.J.L., Bomane, A., Cosson, B., Eyras, E., Rasko, J.E.J., et al. (2017). IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol 18, 51.
Reddy, A.S.N., Marquez, Y., Kalyna, M., and Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683.
Robinson, J.T., Thorvaldsdóttir, H., Wenger, A.M., Zehir, A., and Mesirov, J.P. (2017). Variant review with the integrative genomics viewer. Cancer Res 77, e31–e34.
Song, Q.A., Catlin, N.S., Brad Barbazuk, W., and Li, S. (2019). Computational analysis of alternative splicing in plant genomes. Gene 685, 186–195.
Staiger, D., and Brown, J.W.S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640–3656.
Syed, N.H., Kalyna, M., Marquez, Y., Barta, A., and Brown, J.W.S. (2012). Alternative splicing in plants—coming of age. Trends Plant Sci 17, 616–623.
Szcześniak, M.W., Kabza, M., Pokrzywa, R., Gudyś, A., and Makalowska, I. (2013). Erisdb: a database of plant splice sites and splicing signals. Plant Cell Physiol 54, e10.
Tu, Y.T., Chen, C.Y., Huang, Y.S., Chang, C.H., Yen, M.R., Hsieh, J.W.A., Chen, P.Y., and Wu, K. (2022). HISTONE DEACETYLASE 15 and MOS4-associated complex subunits 3A/3B coregulate intron retention of ABA-responsive genes. Plant Physiol 190, 882–897.
Vicente, J., Mendiondo, G.M., Pauwels, J., Pastor, V., Izquierdo, Y., Naumann, C., Movahedi, M., Rooney, D., Gibbs, D.J., Smart, K., et al. (2019). Distinct branches of the N-end rule pathway modulate the plant immune response. New Phytol 221, 988–1000.
Wang, B.B., and Brendel, V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103, 7175–7180.
Wang, X., Hu, L., Wang, X., Li, N., Xu, C., Gong, L., and Liu, B. (2016). DNA methylation affects gene alternative splicing in plants: an example from rice. Mol Plant 9, 305–307.
Wang, Z., Ji, H., Yuan, B., Wang, S., Su, C., Yao, B., Zhao, H., and Li, X. (2015). ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat Commun 6, 8138.
Wei, G., Liu, K., Shen, T., Shi, J., Liu, B., Han, M., Peng, M., Fu, H., Song, Y., Zhu, J., et al. (2018). Position-specific intron retention is mediated by the histone methyltransferase SDG725. BMC Biol 16, 44.
Yu, Y., Zhang, H., Long, Y., Shu, Y., and Zhai, J. (2022). Plant Public RNA-seq Database: a comprehensive online database for expression analysis of ∼45000 plant public RNA-Seq libraries. Plant Biotechnol J 20, 806–808.
Zhan, X., Qian, B., Cao, F., Wu, W., Yang, L., Guan, Q., Gu, X., Wang, P., Okusolubo, T.A., Dunn, S.L., et al. (2015). An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and aba responses. Nat Commun 6, 8139.
Zhang, H., Zhang, F., Yu, Y., Feng, L., Jia, J., Liu, B., Li, B., Guo, H., and Zhai, J. (2020). A comprehensive online database for exploring ∼20,000 public Arabidopsis RNA-Seq libraries. Mol Plant 13, 1231–1233.
Zhang, R., Calixto, C.P.G., Marquez, Y., Venhuizen, P., Tzioutziou, N.A., Guo, W., Spensley, M., Entizne, J.C., Lewandowska, D., Ten Have, S., et al. (2017). A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucl Acids Res 45, 5061–5073.
Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., Yue, M., Li, Q., Zhang, Y., Xu, Y., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396–411.
Acknowledgements
We thank all the research groups that contributed RNA-seq data to the community, and we regret not being able to cite all the related papers in the main text owing to space constraints. References for all libraries used are listed in PISE ‘All Libraries’. Computation was supported by Center for Computational Science and Engineering at Southern University of Science and Technology. The group of J.Z. was supported by the National Key R&D Program of China (2019YFA0903903), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S172), the Shenzhen Sci-Tech Fund (KYTDPT20181011104005), the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes (2019KSYS006), and the Stable Support Plan Program of Shenzhen Natural Science Fund (20200925153345004). J.J. was supported by the National Natural Science Foundation of China (32100444) and the Shenzhen Fundamental Research Program (JCYJ20210324105202007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Compliance and ethics
The author(s) declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhang, H., Jia, J. & Zhai, J. Plant Intron-Splicing Efficiency Database (PISE): exploring splicing of ∼1,650,000 introns in Arabidopsis, maize, rice, and soybean from ∼57,000 public RNA-seq libraries. Sci. China Life Sci. 66, 602–611 (2023). https://doi.org/10.1007/s11427-022-2193-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-022-2193-3