Abstract
Purpose
The application of fungicides is one of the main strategies to prevent clubroot disease. Currently, numerous studies focus on changes in the soil microbial community at different levels of clubroot disease severity. However, the effects of fungicides on the soil microbial community and causative pathogen, Plasmodiophora brassicae, while preventing clubroot disease remain unclear.
Methods
In this study, we evaluated the control efficacy of three fungicides (fluazinam, metalaxyl-mancozeb, and carbendazim) on clubroot disease of tumorous stem mustard in greenhouse experiment. Uninoculated and Water treatments after inoculation were performed as controls. At three (3 W) and six weeks (6 W) post-inoculation of P. brassicae, soil properties, bacterial composition (sequencing of 16S rRNA genes), and effector gene expression of the pathogen were analyzed. The correlation of these factors with disease index (DI) was explored.
Results
Fluazinam was the most effective in controlling clubroot disease of tumorous stem mustard with a controlled efficacy of 59.81%, and the abundance of P. brassicae in the soil decreased 21.29% after 3 weeks of treatment. Compared with other treatments, twelve out of twenty effector genes showed higher expression in fluazinam 3 W samples. Different fungicides had different effects on soil properties. EC (electrical conductivity), the main factor that positively associated with DI, was significantly lower in fluazinam treatment than the other two fungicide treatments. The application of fungicides, especially carbendazim, significantly reduced bacterial α-diversity and the composition of soil bacteria. Pseudomonas, Microbacterium, and Sphingobacterium (positively correlated with DI) were enriched in Water, metalaxyl-mancozeb, and carbendazim treatments, but were less abundant in fluazinam treatment. Among the three fungicide treatments, DI was significantly negatively correlated with Shannon and Chao 1 indices. Soil properties and the top bacterial genera that positively correlated with DI were influenced to a lesser degree in the fluazinam treatment.
Conclusion
Among three fungicides, fluazinam was the most effective agent with the highest control effects against clubroot disease. The strong virulence of fluazinam against P. brassicae was one of the main reasons for the prevention of clubroot disease, and in addition the alteration of rhizosphere bacterial community by fluazinam to the detriment of P. brassicae infection. Based on our results, EC could be an indicator of the severity of clubroot disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Tumorous stem mustard (Brassica juncea var. tumida) is the raw material of China’s famous pickle, Zhacai, which is mainly grown in the provinces of Chongqing and Zhejiang, and has important economic value. Clubroot disease is one of the main factors affecting the yield and quality of this crop. This disease, caused by Plasmodiophora brassicae, is a devastating disease that threatens the production of cruciferous crops worldwide (Dixon, 2009; Wang et al., 2020). As an obligate biotrophic soil-borne pathogen, P. brassicae resides entirely inside root cells and may survive in the soil for decades, resulting in several barriers to disease control (Rolfe et al., 2016). Currently, chemical fungicides and the selection of disease-resistant varieties are the key strategies against P. brassicae infection in cruciferous crops (Peng et al., 2014; Song et al., 2016; Niemann et al., 2017). In China, a variety of fungicides have been reported to be effective in preventing clubroot disease, but only two, fluazinam and cyazofamid, have been registered (Chai et al., 2014).
The application of fungicides can change soil physicochemical properties and alter the soil microbial community structure (Zhang et al., 2020). In the vegetable-soil ecosystem, with its high microbial abundance and diversity, bacteria play important roles in soil nutrient cycling, regulating the growth of vegetables, and inhibiting soil-borne pathogens (Zhang et al., 2020; Kong et al., 2018). The application of fungicides may directly or indirectly change soil physicochemical properties and soil microbial communities, thus disturbing the stability of the vegetable-soil ecosystem. The effects of fungicide on soil structure and bacterial composition vary under different soil types (Zhang et al., 2020). So far, research on the microbial toxicity of fluazinam to soil is limited. Niemi et al. (2009) showed that fluazinam was highly toxic in microcosms by the luminescent bacteria test; however, further studies on the effects of fluazinam on soil microbial diversity and composition are needed. Soil properties and microbial communities are significantly altered during the infection process of P. brassicae (Lebreton et al., 2019). It is unknown how the application of fungicides in soil contaminated by P. brassicae changes soil properties and microbial communities.
Effector genes, which encode small secreted proteins, are the primary tools for the infection and reproduction of many pathogenic microorganisms (Katagiri, 2018). These genes can target microorganisms, regulating the highly conserved physiological processes of rhizosphere microorganisms, block communication between plants and beneficial microorganisms, or be used to recruit cooperative microorganisms against competitors or for co-colonization of plants (Snelders et al., 2018, 2020). During the infection process, P. brassicae can also secrete a series of effectors to regulate the production of plant hormones, such as cytokinin synthesis, auxin homeostasis, and metabolism of salicylic acid and jasmonic acid, which together stimulate the growth, extension, and differentiation of plant root cells for the formation of galls (Malinowski et al., 2012). However, how effector proteins regulate the soil microbial community requires further research. Whether they can help the pathogen to resist stress and regulate its own growth of the pathogen is still unknown.
Therefore, in this study, three typical fungicides were chosen to evaluate the control efficacy of clubroot disease on tumorous stem mustard and the dynamic change of bacterial communities in the rhizosphere and bulk soils. The analysis of the correlation among soil bacterial communities, clubroot disease severity, soil properties, and expression of effector genes in P. brassicae will help to reveal the molecular ecological mechanism of the clubroot disease development.
2 Materials and methods
2.1 Testing materials
Fluazinam (500 g/L, suspension concentrate, Jiangsu, China), metalaxyl-mancozeb (53%, contains 5% metalaxyl and 48% mancozeb, wettable powder, Shandong, China), and carbendazim (50% wettable powder, Hebei, China) were purchased from Syngenta, Qingdao KYX Chemical Co., Ltd., and Hebei Guanlong Agrochemical Co., Ltd., respectively.
The soil used for greenhouse experiments was collected from 0–20 cm surface soil of an agricultural field where radishes were previously planted in Fuling, Chongqing (29°44′17″ N, 107°12′20″ E). The soil was ground and screened through a 3-mesh sieve, then placed into pots (length × width × height was 12 × 12 × 10 cm).
The vegetable seeds, Fuza No. 2 (Brassica juncea var. tumida Tsen et Lee), were purchased from Chongqing Lvmanyuan Agricultural Science and Technology Co., Ltd. The seeds were treated with 70% ethanol once and then washed three times with sterilized water (1 min for each time). Seeds were sown directly in each pot (in two holes, with 2–3 seeds per hole), and the seedlings were thinned until only two plants remained in each pot at 2 weeks after sowing.
Resting spores of P. brassicae were separated from clubroot galls (stored at − 20 °C) collected from tumorous stem mustard in a field of Baisheng town in Fuling. Galls were washed in tap water to remove soil and other impurities, then washed in sterilized water three times. The inoculation solution was prepared according to Peng et al. (2019), with a slight modification. Galls were chopped up and placed into a tissue crusher with a mass volume ratio of 1:1 to sterilized water. After homogenization, the supernatant was collected by filtering with eight layers of gauze and then centrifuging at 100 × g for 5 min. The concentration of resting spores in the supernatant was counted four times with a hemocytometer, and five meshes were counted each time. Finally, the resting spores were diluted with sterile water to a final concentration of 1 × 108 mL−1 as the inoculation solution.
2.2 Greenhouse experiment
The greenhouse experiment was conducted at Yangtze Normal University (29°45′9″ N, 107°15′24″ E) from May 3 to July 13, 2020 (Fig. 1). The temperature in the greenhouse was kept constant at 24 °C with natural moisture. A total of three fungicide treatments, an uninoculated negative control (Uninoculated), and a water treatment after inoculation (Water) were set up for the study. Each treatment was performed with six replicates containing ten to twelve plants. Inoculation of resting spores and application of fungicides were performed following the protocols of Peng et al. (2019). Three weeks after sowing, plants were inoculated with 2 mL resting spores (1 × 108 mL−1) by pouring the solution around the base of each plant. Uninoculated treatment received 2 mL sterilized water. Twenty-four hours post-inoculation (24 hpi), plants were treated with their assigned fungicide treatments, F-1000, M-500, and C-800, obtained separately by dissolving 1.6 mL fluazinam, 3.2 g metalaxyl-mancozeb, and 2 g carbendazim into 1.6 L tap water (according to the recommended dosage). Ten milliliters of fungicide solution or tap water was poured around the base of each plant as appropriate. Treatment was performed three times at 1, 8, and 18 days post-inoculation (the application times and intervals referred to the instructions). At three weeks (3 W) and six weeks (6 W) post-inoculation, soil samples including the rhizosphere and bulk soil were collected.
Soil samples were collected as described by Kong et al. (2018). Rhizosphere soils (RS) were collected by first shaking off bulk soil. Roots were then agitated in 15 mL phosphate buffer saline (PBS). The suspension was poured into a sterilized centrifuge tube (50 mL). This was repeated three times to obtain a total of 45 mL suspension. The suspension was centrifuged at 8000 × g for 5 min and the supernatant discarded; the resulting sediment pellets were frozen with liquid nitrogen and then stored at − 80 °C for sequencing. Bulk soil (BS) was collected from 10–12 plants and pooled together as a replicate, then divided into two parts. One part was stored at − 80 °C for sequencing. The other part was used to determine soil physicochemical properties.
Plants were harvested at the third and sixth weeks after inoculation. After being washed with PBS solution, the roots were dried with absorbent paper; then, the grade of clubroot disease was determined based on disease grading standards (grade 0: no symptoms; grade 1: small clubs, few galls, mainly on the fibrous root; grade 2: few galls, mainly on the taproot and less on the fibrous roots; grade 3: sever galls on the taproot and the fibrous roots; grade 4: large galls or decayed) (Agarwal et al., 2011). Disease index (DI) was calculated for each treatment at each assessment date using the following formula (Han et al., 2021): DI (%) = ∑ (n × 0 + n × 1 + n × 2 + n × 3 + n × 4) × 100 / (N × 4). Where n is the number of plants in each grade, N is the total number of plants, and 0, 1, 2, 3, and 4 are the symptom severity classes. For each replicate, six roots were pooled together and treated with liquid nitrogen, then stored at − 80 °C for gene expression analysis.
2.3 Physicochemical property of soil samples
All soil sample physicochemical properties were measured by Convinced-Test Company (Nanjing, China). Total nitrogen (TN) (Kjeldahl method, LY/T 1228–2015), total phosphorus (TP) (NaOH alkali fusion-Mo-Sb Anti spectrophotometric method, GB/T 9837–1988), total potassium (TK) (NaOH fusion-flame photometric method, NY/T 87–1988), pH (glass electrode method, NY/T 1121.2–2006), electric conductivity (EC) (electrode method, HJ 802–2016), Ca (HF-HNO3-H2O2 digestion and ICP-AES method), and B (NaOH fusion, ICP-MS method) were detected.
2.4 Amplification and sequencing of 16S rRNA genes
Total soil DNA was extracted with Magnetic Soil and Stool DNA Kit (DP712, TIANGEN BIOTECH CO., LTD, Beijing). The bacterial 16S rRNA genes were amplified with the primer pair 341F (5′-barcode-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) for the V3-V4 region. Amplicons with different barcodes were mixed equally, and TruSeq® DNA PCR-Free Sample Preparation Kit (FC-121–3001, Illumina) was used to construct the DNA library. All 120 soil DNA samples were sequenced by Illumina Novaseq 6000 of CapitalBio Technology (Beijing, China). The sequencing process was performed according to standard protocols, and raw data analysis followed the protocols of Yuan et al. (2018).
After base calling of the original sequencing data, raw data was quality-trimmed with Trimmomatic (Version 0.39). Trailing sequences with quality below 20 were cut out, and sequences with an average quality below 20 were removed with a 15 bp sliding window. Then, fastq-join (Version 1.3.1) and pear were used for joining forward and reverse sequences (minimum overlap was 10 bases and maximum difference was 2 bases). After joining, the primers at both ends were trimmed using Cutadapt (Version 1.18). The sequences without complete primers at both ends were removed. Chimeras were detected and removed by UPARSE-pipeline. Usearch (Version11.0.667) was used to cluster OTU sequences according to 97% similarity. Representative OTU sequences were obtained, and the species annotation conducted with QIIME (RDP Classifier algorithm) based on Silva database. Analysis of soil bacterial α- and β-diversity followed the methods of Tian et al. (2019). Ace, Chao 1, Shannon index, Inv_Simpson index, and rarefaction curves were calculated to evaluate the α-diversity. The relative abundance of soil bacteria was calculated at phylum, class, genus, and OTU levels. For the evaluation of β-diversity, hierarchical cluster dendrograms (Bray–Curtis distance dissimilarities) were constructed according to the composition of the OTUs. Principal coordinate analysis (PCoA), canonical correspondence analysis (CCA), heatmap of relative abundance of the top 40 genera, and Venn diagram were performed using the OmicShare online platform (http://www.omicshare.com/tools). LEfSe (Analysis Effect Size) was performed to identify differentially abundant genera among different treatments (Feng et al., 2017).
2.5 Quantitative PCR (qPCR) of P. brassicae and total bacteria in soil samples
The abundance of bacterial 16S rRNA and P. brassicae in 120 soil samples was tested by qPCR on LightCycler®480 II (Roche, Switzerland). Bacterial 16S rRNA genes were amplified with Eub338F (5′-ACTCCTACGGGAGGCAGCAG-3′)/518R (5′-ATTACCGCGGCTGCTGG-3′) (Fierer et al., 2005), and P. brassicae genes (18S rRNA and ITS1, partial sequence) were amplified with Pb05F (5′-GAACGGGTTCACAGACTAGAT-3′)/05R (5′-GCCCACTGTGTTAATGATCC-3′) (Li et al., 2016). The PCR products were purified with a gel extraction kit (Beijing Suolaibao Technology Co. LTD), cloned into the pMD18-T vector (TaKaRa, Japan), and transformed into DH5α competent cell (Trans5α Chemically Competent Cell, Beijing, China). The positive clones were selected by blue-white screening and sequenced (BGI, Beijing, China). Plasmid DNA in a clone with the correct sequence was extracted by the alkali extraction method and then used for building a standard curve (Wei et al., 2018; Wallenhammar et al., 2012). Copy numbers were log10-transformed to normalized values (Wei et al., 2018). The qPCR reaction mixture contained 5 μL of Master Mix (Eva Green 2 × qPCR Mix-no Dye, ABM, Canada), 0.5 μL of each primer, 0.5 μL of template DNA, and 3.5μL of nuclease-free water. The PCR cycling conditions were as follows: 95 °C for 5 min, 45 cycles of 95 °C for 15 s, 60 °C for 15 s, 72 °C for 35 s, the melting curve obtained by 95 °C continuous. Each sample was amplified with three technical replicates. Standard curve samples were obtained through ten times gradient dilution and conducted with three technical replicates for each concentration.
2.6 Gene expression of P. brassicae
Total RNA of galls was extracted with RNAsimple Total RNA Kit following the manufacturer’s instructions (Tiangen Biotech CO., LTD). The RNA concentration and purity were determined by Nanodrop One and then stored at − 80 °C. Genomic DNA was removed, and cDNA was synthesized using 5 × All-In-One RT MasterMix (with AccuRT Genomic DNA Removal Kit) (Applied Biological Materials Inc, ABM). 2 μg RNA was reverse-transcribed according to the reagent composition in the instructions. The obtained cDNA was diluted ten times and used as a qPCR template. The expression of 20 candidate effector genes was tested (Table S1). Pbactin (AAR88383) was used as the reference gene (Feng et al., 2013). All primers used in this protocol are listed in Additional file 1. Gene expression data were analyzed by the 2−ΔCT method (Manini et al., 2011). A minimum of six plants were pooled into one sample for each treatment, and each sample was run with three technical replicates.
2.7 Statistical analysis
IBM SPSS Statistics 22 was used for general statistical analysis. Data of disease severity, the abundance of P. brassicae and soil bacteria, soil properties, and α-diversity indices were analyzed by one-way analysis of variance (ANOVA, P < 0.05). The comparison of two sets of data (3 W and 6 W, RS and BS) was completed by Student’s t-test. MRPP (multi-response permutation procedure), ANOSIM (analysis of similarities), and PERMANOVA (permutational analysis of variance) were conducted to assess the significant differences in bacterial communities among different treatments (Feng et al., 2017). The relationship between soil properties, gene expression, and bacterial communities was analyzed by Mantel tests (based on Bray–Curtis) (Feng et al., 2017). Pearson’s correlation coefficient was employed to correlate the soil properties with DI and α-diversity by SPSS 22. Network analysis was based on MENA online platform (Deng et al., 2012), and data visualization was performed with Cytoscape software. General figures were drawn with SigmaPlot 10.0.
3 Results
3.1 Effects of fungicides on clubroot disease of tumorous stem mustard
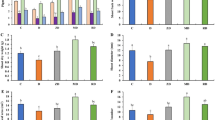
Compared with Water treatment, the three fungicides inhibited the occurrence and severity of clubroot disease to a certain extent after 3 and 6 weeks of treatment (Fig. 2 A and B). The F-1000 treatment showed the highest control efficacy (59.81%), with the lowest disease incidence and DI, significantly lower than the Water control and the other two fungicide treatments after 6 weeks of treatment. There were no significant differences between M-500 (15.95%) and C-800 (29.80%) treatments in the control effect, but the disease incidence and DI were significantly lower than that of Water treatment (Fig. 2B). Three weeks after inoculation, the abundance of P. brassicae in rhizosphere soil (RS) was consistent with the severity of clubroot disease. Six weeks after inoculation, the abundance of P. brassicae increased in RS, and the Water and M-500 treatments were significantly higher than C-800 and F-1000 treatments. There was no significant difference in bulk soil between the two sampling time points (Fig. 2C). Expression of candidate effector genes (Figure S1) showed that 3 W samples of F-1000 formed a separate cluster, in which twelve of the twenty genes displayed the highest expression level among all samples.
Clubroot disease severity and abundance of P. brassicae in soil under different fungicide treatments. (A) Phenotype of clubroot in different treatments. (B) Disease incidence and disease index in different treatments. (C) The abundance of P. brassicae in soil. All data were calculated based on six replicates, and bars indicate standard errors (SE). Different letters indicated significant differences among the treatments by one-way ANOVA test at P < 0.05 (LSD in B, and Duncan in C). M-500, 500-fold dilution of metalaxyl-mancozeb; C-800, 800-fold dilution of carbendazim; F-1000, 1000-fold dilution of fluazinam; Water, water treatment after inoculation; Uninoculated, uninoculated treatment. RS, rhizosphere soil; BS, bulk soil; 3 W, three weeks after inoculation; 6 W, 6 weeks after inoculation
3.2 Inoculation of P. brassicae and the application of fungicides affect the physicochemical properties of soils
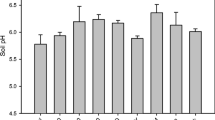
The application of different fungicides resulted in various differences in soil physicochemical properties (Fig. 3 and Table S2). Three weeks after inoculation (3 W), compared with the healthy soil (Uninoculated treatment), TN and EC in the Water treatment significantly increased, while Ca significantly decreased. Six weeks after inoculation (6 W), TK, EC, and B were significantly increased in the Water treatment compared with the Uninoculated treatment. However, except for TN and pH, which significantly increased in F-1000 treatment compared with Uninoculated soil, the F-1000 treatment (with the lowest disease index) offset the changes in soil properties altered by the inoculation of P. brassicae, and soil properties tend to be consistent with that of healthy soil, particularly EC. A different trend of soil property changes was observed in the M-500 and C-800 treatments, with EC significantly increasing in those soil samples.
Effects of different treatments on physicochemical properties of soils. All data are represented with mean ± SE. Different letters indicated significant differences among different treatments at P < 0.05 by one-way ANOVA test. Asterisks indicated significant differences between 3 and 6 W samples at each treatment based on Student’s t-test (*P < 0.05, **P < 0.01). M-500, 500-fold dilution of metalaxyl-mancozeb; C-800, 800-fold dilution of carbendazim; F-1000, 1000-fold dilution of fluazinam; Water, water treatment after inoculation; Uninoculated, uninoculated treatment. 3 W, 3 weeks after inoculation; 6 W, 6 weeks after inoculation. TN, total nitrogen; TP, total phosphorus; TK, total potassium; pH, potential of hydrogen; EC, electric conductivity; Ca, calcium; B, boron
3.3 The application of fungicides led to a typical reduction of α-diversity
In this study, a total of 8,624,010 raw bacterial sequences were obtained from 120 soil samples through Illumina Novaseq 6000, of which 8,602,592 high-quality sequences were retained after quality control steps. These sequences were clustered into 1657 to 4223 OTUs per sample. The rarefaction curves demonstrated that the sequencing depths were appropriate (Figure S2). The α-diversity indices (Ace, Chao 1, Shannon index, Inv_Simpson index) significantly decreased in the 6 W rhizosphere soil samples, especially in the fungicide treatment samples. C-800 samples showed the lowest α-diversity among all treatments (Fig. 4) but had the highest abundance of total bacteria in the 6 W RS samples (Figure S3). Uninoculated group samples showed little change in α-diversity, and there were no significant differences between all samples in this group.
The α-diversity of soil bacterial communities. All data are represented with mean ± SE. Different lowercase letters indicated significant differences among different treatments, and different capital letters indicated significant differences in one treatment at P < 0.05 (one-way ANOVA test). RS, rhizosphere soil; BS, bulk soil. M-500, 500-fold dilution of metalaxyl-mancozeb; C-800, 800-fold dilution of carbendazim; F-1000, 1000-fold dilution of fluazinam; Water, water treatment after inoculation; Uninoculated, uninoculated treatment. 3 W, 3 weeks after inoculation; 6 W, 6 weeks after inoculation
3.4 Effects of different treatments on bacterial community composition
Principal coordinate analysis (PCoA) and dissimilarity analysis (MRPP, ANOSIM, and PERMANOVA) demonstrated that inoculation of P. brassicae and application of fungicides significantly altered bacterial community composition (Fig. 5, Additional file 2). The two axes explained a total of 52.15% and 37.92% of the variation in the RS and BS samples, respectively. The dominant phyla were Proteobacteria (37.89–78.40%), Acidobacteria (1.90–18.80%), Actinobacteria (4.75–18.11%), Chloroflexi (0.77–9.20%), Bacteroidetes (1.84–9.28%), and Gemmatimonadetes (0.52–5.27%) in all soil samples, accounting for 80.96–94.71% of relative abundance (Table S3). The changes in bacterial composition of BS samples were less than that of RS samples (Figure S4). After inoculation and application of fungicides, Proteobacteria and Bacterioidetes were significantly enriched in 6 W RS samples, while Acidobacteria, Chloroflexi, and Gemmatimonadetes showed the opposite trend (Fig. 6).
Principal coordinate analysis (PCoA) of bacteria communities. The analysis was conducted based on the Bray–Curtis distance. RS, rhizosphere soil; BS, bulk soil; M-500, 500-fold dilution of metalaxyl-mancozeb; C-800, 800-fold dilution of carbendazim; F-1000, 1000-fold dilution of fluazinam; Water, water treatment after inoculation; Uninoculated, uninoculated treatment. 3 W, 3 weeks after inoculation; 6 W, 6 weeks after inoculation
Differences in the relative abundance of dominant phyla in RS samples. Different letters indicated significant differences of that phylum in different treatments at the P < 0.05 level (one-way ANOVA, LSD). RS, rhizosphere soil; M-500, 500-fold dilution of metalaxyl-mancozeb; C-800, 800-fold dilution of carbendazim; F-1000, 1000-fold dilution of fluazinam; Water, water treatment after inoculation; Uninoculated, uninoculated treatment. RS, rhizosphere soil; BS, bulk soil; 3 W, 3 weeks after inoculation; 6 W, 6 weeks after inoculation
The bacterial community composition at the genus level changed markedly after inoculation, especially in the 6 W samples, which showed clear differences with Uninoculated soil (Fig. 7). Cluster analysis of the genera showed that these bacteria could be divided into three groups according to their relative abundance in different samples, marked as B1 to B3. B1 genera (such as Enterobacter, Comamonas, Brevundimonas, and Delftia) were greatly enriched in the 6 W samples of inoculation and fungicide treatments. B3 genera (such as Acidothermus, Bryobacter, and Gaiella) were greatly decreased in the 6 W samples of inoculation and fungicide treatments. The change trends of B2 genera were irregular, with some showing an increased relative abundance in different treatments. After the application of fungicides, the 6 W samples of C-800 treatment showed the highest reduction in these genera.
Relative abundance of top 40 genera in RS samples. RS, rhizosphere soil; M-500, 500-fold dilution of metalaxyl-mancozeb; C-800, 800-fold dilution of carbendazim; F-1000, 1000-fold dilution of fluazinam; Water, water treatment after inoculation; Uninoculated, uninoculated treatment. 3 W, 3 weeks after inoculation; 6 W, 6 weeks after inoculation. B1, B2, and B3 were three clusters divided by clustering
Differentially abundant genera were found among the RS bacteria communities (3 W and 6 W) based on discriminant analysis effect size (LEfSe) (Figure S5). The Uninoculated, F-1000, and C-800 treatments contained the greatest, the second greatest, and the least number of genera, respectively. The Uninoculated, Water, and F-1000 treatments showed a greater number of these genera in the 6 W samples than in the 3 W samples. The opposite was observed in M-500 and C-800. There were 9, 2, 0, 1, and 8 common differentially abundant genera in the 3 W and 6 W samples of Uninoculated, Water, M-500, C-800, and F-1000 treatments, respectively.
3.5 Correlation between disease severity, gene expression, soil properties, and bacterial communities
Pearson correlation analysis (Table S4 and S5), Mantel test (Table S6), and network analysis (Fig. 8) showed that the soil properties correlated with bacterial communities were different after application of the different fungicides. The correlation among α-diversity, soil properties, and disease index was the most complex in the C-800 treatment, followed by F-1000 treatment, and the simplest in the M-500 treatment. Among the three fungicide treatments, DI was significantly negatively correlated with the Shannon and Chao 1 indices and significantly positively correlated with EC and TK. Among the seven soil properties, only TP was significantly negatively correlated with α-diversity. Canonical correspondence analysis (CCA) results showed that soil physicochemical factors explained 64.87% and 62.56% of variation in the RS and BS bacterial communities, respectively (Figure S6).
Correlation network of α-diversity, disease index (DI), and soil properties. Green, purple, and red circles represented α-diversity indices, soil properties, and DI, respectively. The green and red lines indicated significant negative and positive correlations, respectively. Correlation between DI and α-diversity, DI and soil properties, α-diversity, and soil properties referred to Table S4 and S5. The network was conducted with Cytoscape software. TP, total phosphorus; TK, total potassium; pH, potential of hydrogen; EC, electric conductivity; Ca, calcium
Comamonas, Delftia, Enterobacter, Microbacterium, Pectobacterium, Pseudomonas, and Sphingobacterium, which belonged to group B1, were significantly positively correlated with DI (Additional file 4), of which, three genera were differentially abundant genera in Water (Comamonas, Delftia, and Enterobacter) and C-800 treatments (Microbacterium, Pseudomonas, and Sphingobacterium), respectively. A single genus (Pectobacterium) was differentially abundant in the M-500 treatment (Figure S6). Another twenty-five genera in B2 and B3 were significantly negatively correlated with DI (Additional file 4), among which nineteen and three genera were differentially abundant genera in the Uninoculated and F-1000 treatments, respectively (Figure S5). The correlation analysis between the top 40 genera (Additional file 3) and the expression of candidate effector genes (Figure S7) showed that Pectobacterium was significantly positively correlated with the expression of nine effector genes, indicating that this genus may be greatly affected by the effectors. Pb9670 and Pb4539 were negatively correlated with seven and nine genera, respectively, suggesting that these two effectors may act on various bacteria and may be the core effectors for the regulation of rhizosphere microorganisms.
4 Discussion
4.1 Fluazinam is the most effective fungicide to control clubroot disease on tumorous stem mustard in Fuling
In the present study, it was found that the control efficacy of fluazinam was 59.81%, which was significantly higher than the control effect of carbendazim and metalaxyl-mancozeb (Fig. 2B). Suzuki et al. (1995) demonstrated that fluazinam could inhibit the germination of resting spores and the root hair infection stage, resulting in the inhibition of club formation. However, our results proved that the application of fluazinam could reduce the abundance of P. brassicae in the soil. In addition, soil properties in fluazinam treatment were influenced to a lesser degree when compared with the healthy soils. Enterobacter, Microbacterium, and Pectobacterium etc. (significantly positive correlation with DI) were less enriched in fluazinam treatment. Therefore, beyond its direct influence on clubroot pathogens, fluazinam likely also controls clubroot disease by changing the bacterial community of the soil.
We further analyzed the effect of fluazinam on candidate effector genes of P. brassicae. Effectors, as a group of secretory proteins, are critical for the successful infection of many fungi, oomycetes, bacteria, and nematodes (Chen et al., 2019). Numerous studies have been conducted to explore their functions in the infection process of pathogens, but little is known about their role during fungicide treatment. Our data showed that twelve out of twenty candidate effector genes were highly expressed in F-1000 3 W samples which showed the lowest DI (Figure S1), indicating that these effectors were potentially stimulated by fluazinam. Under the stress of fluazinam, P. brassicae may regulate candidate effector genes to complete the infection process. The correlation analysis between the top 40 genera and the expression of candidate effector genes (Figure S7) showed that Pectobacterium was significantly positively correlated with the expression of nine effector genes, indicating that this genus may be greatly affected by the effectors. F-1000 treatment showed an increase of P. brassicae in the soil from 3 to 6 W (Fig. 2C), resulting in no significant difference between F-1000 and Water treatment for the estimated amount of P. brassicae biomass in 6 W RS samples. This may indicate that there is a short cycle during the infection to increase the amount of P. brassicae in the soil rapidly (Buczacki and Clay, 1984), or secondary zoospores released from root hair or cortex cells resulted in this phenomenon (Liu et al., 2020). Whether these effectors were related to the increase of P. brassicae in soil requires further exploration.
4.2 EC was positively correlated with DI of clubroot disease, and the correlation was the highest among all soil properties
At present, there have been few reports on the variation of EC in soil during the infection process of P. brassicae on Cruciferae plants. In our study, EC was significantly positively related to DI, and had the highest correlation value (Table S4, Fig. 8). Li et al. (2018) suggesting that EC could be a new tool for monitoring soil-borne disease. Arif et al. (2021) found that the relative electrical conductivity of roots inoculated with P. brassicae was 2.85 times greater than that of the control, which may be due to the increased membrane permeability and cell membrane damage. In our study, the F-1000 treatment (with the least disease) showed a lower EC value than the other inoculated treatments (Table S2). These results might indicate that EC could be an indicator of the severity of clubroot disease.
Other factors affecting the occurrence of clubroot disease in soil, including soil pH, Ca, B, soil enzyme activity, temperature, water, organic matter, soil type, soil fertility, and abundance of P. brassicae, were also reported (Bhering et al., 2020). In this study, the mean value of soil pH under different treatments varied from 5.4 to 6.2 (Fig. 3). The pH was significantly higher in the M-500 (with severe disease) and F-1000 (with the least disease) treatments, indicating that the correlation between pH (5.4–6.2) and the severity of clubroot disease was weak, consistent with the report of Gossen et al. (2013). Thus, the significant negative correlation between pH and DI in the F-1000 treatment may be caused by other factors. B and Ca are also important factors influencing the infection of P. brassicae, and their application to soil could inhibit infection to some extent (Dixon, 1996; Webster and Dixon, 2009). In the current study, the contents of B and Ca in soil under different treatments were detected, and it was determined that the content of B in the soil at 6 W (water) after inoculation was significantly higher than that in the Uninoculated group (Fig. 3). It was speculated that the B might come from plants or soil organic matter (Parks and White, 1952; Yermiyahu et al., 1988; Hayes and Reid, 2004). Additionally, the content of B in soil treated with F-1000 at 3 W was significantly higher than that at 6 W (Fig. 3). Whether these changes were related to plant defense requires further study.
4.3 The variation of the main bacterial community in soil treated with fungicides and the infection of P. brassicae
Soil bacteria, as an important part of the soil microbial community, are of great significance in maintaining soil structure and function, and microbial diversity is positively correlated with the suppression of disease (Saraiva et al., 2020). The development of clubroot disease is determined by the soil environment, including its properties and biological characteristics (Bhering et al., 2020). The interaction of soil properties, soil microbial diversity, and clubroot development is complex and dynamic, but no cause-and-effect relationship between bacterial composition and clubroot severity has been found (Bhering et al., 2020). Meanwhile, other studies have summarized the characteristics of bacterial composition in soils with severe clubroot disease (Lebreton et al., 2019; Saraiva et al., 2020). In our study, we found that Proteobacteria was the dominant phylum in all soil samples, which is consistent with previous studies (Wang et al., 2020; Lebreton et al., 2019). Numerous studies have indicated that Proteobacteria and Acidobacteria may act as bioindicators of soil suppression to phytopathogens (Lee et al., 2017; Liu et al., 2016; Xiong et al., 2017; Shen et al., 2015). In our study, we found that Proteobacteria and Bacteroidetes were positively correlated with soil samples experiencing severe clubroot disease. At the same time, Acidobacteria, Actinobacteria, Chloroflexi, and Gemmatimonadetes were positively correlated with soil samples with less clubroot disease (3 W and Uninoculated samples) (Figure S8). Therefore, the abundance of Proteobacteria may be stimulated by pathogens in the soil, or that these bacteria are less sensitive to fungicides. That Acidobacteria were positively correlated with soil samples experiencing lower clubroot severity is consistent with the results of Saraiva et al. (2020).
Nitrospira and Bradyrhizobium, as shown by Saraiva et al. (2020) to be more abundant in the soils containing plants with lower clubroot severity, were significantly negatively correlated with DI in our study (Additional file 4). Similar to Lebreton et al. (2019), the rhizosphere soil bacterial communities were disrupted to a greater degree by the invasion of P. brassicae during the latter part of the incubation (6 W) (Figs. 5, 7, S7, and S9). Flavisolibacter, Streptomyces, and Bacillus showed low abundance in 6 W RS soil samples with severe clubroot disease, consistent with Lebreton et al. (2019). The application of fungicides usually significantly affects bacterial community composition in soil. Sample cluster analysis of the top forty genera showed that the 6 W samples of Water, M-500, and C-800 formed a cluster. In contrast, all (3 W and 6 W) samples of Uninoculated treatment, and 3 W samples of Water, and M-500 were another cluster (Fig. 7). These results may indicate that the dominant bacterial communities in M-500 treatment may be greatly affected by the infection of P. brassicae. The influence of M-500 on these bacteria still followed a changing trend during the infection process of P. brassicae.
4.4 Sensitive bacteria are markedly altered by the application of fungicides
According to our results (Fig. 4), fungicides significantly altered bacterial diversity, especially the carbendazim (C-800) treatment. The carbendazim treatment showed the greatest influence on α-diversity, consistent with the research of Wang et al. (2012). You et al. (2016) reported that the fungicide metalaxyl-mancozeb only slightly decreased the bacterial community Shannon–Wiener index but substantially decreased the Margalef richness index. In this study, some bacterial species and genera were eliminated by the application of fungicides (Figure S9). We screened for genera and species present in Uninoculated and Water treatments but absent in the different fungicide treatments. According to the results, there were nine genera and ten species that were not detected in all three fungicide treatments. The C-800 treatment contained the most (twenty-five) undetected species, including Bacillus polymachus, which displays a broad range of antibacterial activity (Nguyen and Kim, 2015). The undetected species and genera in fungicide treatments are listed in Additional file 5.
5 Conclusions
The control efficacy, soil properties, bacterial community, and effector gene expression were markedly different under the different fungicide treatments. As the most effective fungicide, fluazinam had the strongest effect upon P. brassicae, significantly reducing the amount of P. brassicae in the soil. Another reason for its intense control effect might be the altered composition of rhizosphere soil bacterial community. Genera that displayed a significant positive correlation with DI were mainly differentially abundant genera in C-800 (800-fold dilution of carbendazim) and Water treatments. In contrast, the genera with significant negative correlation with DI were mainly differentially abundant genera in Uninoculated and F-1000 (1000-fold dilution of fluazinam) treatments. Based on our results, EC could be used as an indicator of the severity level of clubroot disease, but the specific criteria need to be further clarified. Whether the stimulation of fluazinam on effector genes leads to the rapid reproduction in the later period should be considered in future studies. In addition, future work should also involve the effects of effector genes on the soil bacterial community and the pathogenicity of P. brassicae.
Data availability
Sequencing raw data is available online: http://www.ncbi.nlm.nih.gov/genbank/, PRJNA688622.
References
Agarwal A, Kaul V, Faggian R, Rookes JE, Ludwig-Müller J, Cahill DM (2011) Analysis of global host gene expression during the primary phase of the Arabidopsis thaliana-Plasmodiophora brassicae interaction. Funct Plant Biol 38:462–478. https://doi.org/10.1071/FP11026
Arif S, Liaquat F, Yang S, Shah IH, Zhao L, Xiong X, Garcia D, Zhang Y (2021) Exogenous inoculation of endophytic bacterium Bacillus cereus suppresses clubroot (Plasmodiophora brassicae) occurrence in pak choi (Brassica campestris sp. chinensis L.). Planta 253: 25.
Bhering AS, Carmo MGF, Coelho IS, Lima ESA, de Carvalho CF, Saraiva ALRF, Passos SR, Sobrinho NMBA (2020) Soil management in a mountain agroecosystem and clubroot disease. Plant Pathol 69:302–309. https://doi.org/10.1111/ppa.13123
Buczacki ST, Clay CM (1984) Some observations on secondary zoospore development in Plasmodiophora brassicae. Trans Br Mycol Soc 82:339–382. https://doi.org/10.1016/S0007-1536(84)80079-0
Bulman S, Richter F, Marschollek S, Benade F, Jülke S, Ludwig-Müller J (2018) Arabidopsis thaliana expressing PbBSMT, a gene encoding a SABATH-type methyltransferase from the plant pathogenic protist Plasmodiophora brassicae, show leaf chlorosis and altered host susceptibility. Plant Biol 21(S1):120–130. https://doi.org/10.1111/plb.12728
Chai AL, Xie XW, Shi YX, Li BJ (2014) Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can J Plant Pathol 36:142–153. https://doi.org/10.1080/07060661.2013.868829
Chen W, Yan Li, Yan R, Xu L, Ren L, Liu F, Zeng L, Yang H, Chi P, Wang X, Chen K, Ma D, Fang X (2019) Identification and characterization of Plasmodiophora brassicae primary infection effector candidates that suppress or induce cell death in host and nonhost plants. Phytopathology 109:1689–1697. https://doi.org/10.1094/PHYTO-02-19-0039-R
Deng Y, Jiang YH, Yang Y, He Z, Luo F, Zhou J (2012) Molecular ecological network analyses. BMC Bioinformatics 13:113
Dixon GR (1996) Repression of the morphogenesis of Plasmodiophora brassicae Wor. By boron-a review. Acta Hort 407: 393–401. https://doi.org/10.17660/ActaHortic.1996.407.50.
Dixon GR (2009) The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul 28:194–202. https://doi.org/10.1007/s00344-009-9090-y
Djavaheri M, Ma L, Klessig DF, Mithöfer A, Gropp G, Borhan H (2019) Mimicking the host regulation of salicylic acid: a virulence strategy by the clubroot pathogen Plasmodiophora brassicae. MPMI 32(3):296–305. https://doi.org/10.1094/MPMI-07-18-0192-R
Feng J, Hwang SF, Strelkov SE (2013) Assessment of gene expression profiles in primary and secondary zoospores of Plasmodiophora brassicae by dot blot and real-time PCR. Microbiol Res 168:518–524
Feng K, Zhang Z, Cai W, Liu W, Xu M, Yin H, Wang A, He Z, Deng Y (2017) Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol Ecol 26:6170–6182. https://doi.org/10.1111/mec.14356
Fierer N, Jackson JA, Vilgalys R, Jackson RB (2005) Assement of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microb 71:4117–4120. https://doi.org/10.1128/AEM.71.7.4117-4120.2005
Gossen BD, Kasinathan H, Cao T, Manolii VP, Strelkov SE, Hwang SF, McDonald MR (2013) Interaction of pH and temperature affect infection and symptom development of Plasmodiophora brassicae in canola. Can J Plant Pathol 35:294–303. https://doi.org/10.1080/07060661.2013.804882
Han X, Yin J, Ullah I, Luo E, Yue Y (2021) Plasmodiophora brassicae in Yunnan and its resistant sources in Chinese cabbage. Intl J Agric Biol 25:805–812
Hayes JE, Reid RJ (2004) Boron tolerance in barley is mediated by efflux of B from the roots. Plant Physiol 136:3376–3382. https://doi.org/10.1104/pp.103.037028
Katagiri F (2018) Review: Plant immune signaling from a network perspective. Plant Sci 276:14–21
Kong X, Jin D, Jin S, Wang Z, Yin H, Xu M, Deng Y (2018) Responses of bacterial community to dibutyl phthalate pollution in a soil-vegetable ecosystem. J Hazard Mater 353:142–150. https://doi.org/10.1016/j.jhazmat.2018.04.015
Lebreton L, Guillerm-Erckelboudt A, Gazengel K, Linglin J, Ourry M, Glory P, Sarniguet A, Daval S, Manzanares-Dauleux M·J, Mougel C, (2019) Temporal dynamics of bacterial and fungal communities during the infection of Brassica rapa roots by the protist Plasmodiophora brassicae. PLoS ONE 14:e0204195. https://doi.org/10.1371/journal.pone.0204195
Lee CG, Lida T, Inoue Y, Muramoto Y, Watanabe H, Nakaho K, Ohkuma M (2017) Prokaryotic communities at different depths between soils with and without tomato bacterial wilt but pathogen-present in a single greenhouse. Microbes Environ 32:118–124. https://doi.org/10.1264/jsme2.ME16136
Li M, Zhou H, Liu Y, Liu F, Yang J, Ji G (2016) Detection of Plasmodiophora brassicae with real-time quantitative PCR in Yunnan province. Journal of Yunnan Agricultural University (natural Science) 31:43–48 ((In Chinese))
Li Z, Deng Z, Chen S, Yang H, Zheng Y, Dai L, Zhang F, Wang S, Hu SJ (2018) Contrasting physical and biochemical properties of orchard soils suppressive and conducive to Fusarium wilt of banana. Soil Use Manage 34:154–162. https://doi.org/10.1111/sum.12390
Liu L, Qin L, Zhou Z, Hendriks W, Liu S, Wei Y (2020) Refining the life cycle of Plasmodiophora brassicae. Phytopathology 110:1704–1712. https://doi.org/10.1094/PHYTO-02-20-0029-R
Liu X, Zhang S, Jiang Q, Bai Y, Shen G, Li S, Ding W (2016) Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci Rep-UK 6:e36773. https://doi.org/10.1038/srep36773
Ludwig-Müller J, Jülke S, Geiß K et al (2015) A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol Plant Pathol 16(4):349–364
Malinowski R, Smith JA, Fleming AJ et al (2012) Gall formation in clubroot-infected Arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J 71(2):226–238
Manini TM, Vincent KR, Leeuwenburgh CL, Lees HA, Kavazis AN, Borst SE, Clark BC (2011) Myogenic and proteolytic mRNA expression following blood flow restricted exercise. Acta Physiol (oxf) 201:255–263. https://doi.org/10.1111/j.1748-1716.2010.02172.x
Nguyen TM, Kim J (2015) Bacillus polymachus sp. Nov. with a broad range of antibacterial activity, isolated from forest topsoil samples by using a modified culture method. Int J Syst Evol Micr 65:704–709. https://doi.org/10.1099/ijs.0.070326-0
Niemann J, Kaczmarek J, Ksiazczyk T, Wojciechowski A, Jedryczka M (2017) Chinese cabbage (Brassica rapa spp. Pekinensis)-a valuable source of resistance to clubroot (Plasmodiophora brassicae). Eur J Plant Pathol 147:181–198. https://doi.org/10.1007/s10658-016-0991-x
Niemi RM, Heiskanen I, Ahtiainen JH, Rahkonen A, Mantykoski K, Welling L, Laitinen P, Ruuttunen P (2009) Microbial toxicity and impacts on soil enzyme activities of pesticides used in potato cultivation. Appl Soil Ecol 41:293–304. https://doi.org/10.1016/j.apsoil.2008.12.002
Parks WL, White JL (1952) Boron retention by clay and humus systems saturated with various cations. Soil Sci Soc Amer Proc 16:298–300. https://doi.org/10.2136/sssaj1952.036159950016000030018x
Peng G, Lahlali R, Hwang SF, Pageau D, Hynes RK, McDonald MR, Gossen BD, Strelkov SE (2014) Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can J Plant Pathol 36:99–112. https://doi.org/10.1080/07060661.2013.860398
Peng Y, Gossen BD, Huang Y, Al-Daoud F, McDonald MR (2019) Development of Plasmodiophora brassicae in the root cortex of cabbage over time. Eur J Plant Pathol 154(3):727–737. https://doi.org/10.1007/s10658-019-01696-0
Pérez-López E, Hossain M, Tu J, Waldner M, Todd CD, Kusalik AJ, Wei Y, Bonham-Smith PC (2020) Transcriptome analysis identifies Plasmodiophora brassicae secondary infection effector candidates. J Eukaryot Microbiol 67:337–351. https://doi.org/10.1111/jeu.12784
Rolfe RA, Strelkov SE, Links MG, Clarke WE, Robinson SJ, Djavaheri M, Malinowski R, Haddadi P, Kagale S, Parkin IAP, Taheri A, Borhan MH (2016) The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genomics 17:272. https://doi.org/10.1186/s12864-016-2597-2
Saraiva A, Bhering AS, Carmo M, Andreote FD, Dias A, Coelho IS (2020) Bacterial composition in brassica-cultivated soils with low and high severity of clubroot. J Phytopathol 168:613–619. https://doi.org/10.1111/jph.12941
Schwelm A, Fogelqvist J, Knaust A, Jülke S, Lilja T, Bonilla-Rosso G, Karlsson M, Shevchenko A, Dhandapani V, Choi SR, Kim HG, Park JY, Lim YP, Ludwig-Müller J, Dixelius C (2015) The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci Rep-UK 5:11153. https://doi.org/10.1038/srep11153
Shen Z, Ruan Y, Xue C, Zhong S, Li R, Shen Q (2015) Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil 393:21–33
Snelders NC, Kettles GJ, Rudd JJ et al (2018) Plant pathogen effector proteins as manipulators of host microbiomes? Mol Plant Pathol 19(2):257–259
Snelders NC, Rovenich H, Petti GC et al (2020) Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat Plants 6(11):1365–1374
Song T, Chu M, Lahlali R, Yu F, Peng G (2016) Shotgun label-free proteomic analysis of clubroot (Plasmodiophora brassicae) resistance conferred by the gene Rcr1 in Brassica rapa. Front Plant Sci 7:1013. https://doi.org/10.1007/s11104-015-2474-9.
Suzuki K, Sugimoto K, Hayashi H, Komyoji T (1995) Biological mode of action of fluazinam, a new fungicide, for Chinese cabbage clubroot. Ann Phytopathol Soc Jpn 61:395–398. https://doi.org/10.3186/jjphytopath.61.395
Tian X, Wang D, Mao Z, Pan L, Liao J, Cai Z (2019) Infection of Plasmodiophora brassicae changes the fungal endophyte community of tumourous stem mustard roots as revealed by high-throughput sequencing and culture dependent methods. PLoS ONE 14(6):e0214975. https://doi.org/10.1371/journal.pone.0214975
Wang D, Sun T, Zhao S, Pan L, Liu H, Tian X (2020) Physiological change alters endophytic bacterial community in clubroot of tumorous stem mustard infected by Plasmodiophora brassicae. BMC Microbiol 20:244. https://doi.org/10.21203/rs.3.rs-28394/v1.
Wang X, Song M, Wang Y, Gao C, Zhang Q, Chu X, Fang H, Yu Y (2012) Response of soil bacterial community to repeated applications of carbendazim. Ecotoxicol Environ Saf 75:33–39. https://doi.org/10.1016/j.ecoenv.2011.08.014
Webster MA, Dixon GR (2009) Boron, pH and inoculum concentration influencing colonization by Plasmodiophora brassicae. Mycol Res 95:74–79
Wallenhammar AC, Almquist C, Söderström M, Jonsson A (2012) In-field distribution of Plasmodiophora brassicae measured using quantitative real-time PCR. Plant Pathol 61:16–28
Wei W, Yang M, Liu Y, Huang H, Ye C, Zheng J, Guo C, Hao M, He X, Zhu S (2018) Fertilizer N application rate impacts plant-soil feedback in a sanqi production system. Sci Total Environ 633:796–807
Xiong WU, Li R, Ren YI, Liu C, Zhao Q, Wu H, Shen Q (2017) Distinct roles for soil fungal and bacterial communities associated with suppression of vanilla Fusarium wilt disease. Soil Biol Biochem 107:198–207. https://doi.org/10.1016/j.soilbio.2017.01.010
Yermiyahu U, Keren R, Chen Y (1988) Boron sorption on composted organic matter. Soil Sci Soc Am J 52:1309–1313. https://doi.org/10.2136/sssaj1988.03615995005200050018x
You C, Zhang C, Kong F, Feng C, Wang J (2016) Comparison of the effects of biocontrol agent Bacillus subtilis and fungicide metalaxyl-mancozeb on bacterial communities in tobacco rhizospheric soil. Ecol Eng 91:119–125. https://doi.org/10.1016/j.ecoleng.2016.02.011
Yuan X, Long Y, Ji Z, Gao J, Fu T, Yan M, Zhang L, Su H, Zhang W, Wen X, Pu Z, Chen H, Wang Y, Gu X, Yan B, Kaliannan K, Shao Z (2018) Green tea liquid consumption alters the human intestinal and oral microbiome. Mol Nutr Food Res 62:1800178. https://doi.org/10.1002/mnfr.201800178
Zhang H, Song J, Zhang Z, Zhang Q, Chen S, Mei J, Yu Y, Fang H (2020) Exposure to fungicide difenoconazole reduces the soil bacterial community diversity and the co-occurrence network complexity. J Hazard Mater 405:124208.
Acknowledgements
We would like to thank the staff of Yangtze Normal University for providing the research platform and logistics services.
Funding
This research was funded by Youth Program of National Natural Science Foundation of China (31800311) and Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJQN201801431).
Author information
Authors and Affiliations
Contributions
JJL designed and performed all experiments and writing the manuscript draft. LZ, LZW, XDS, HY, SHT, LT, and XL gave strong support in acquisition of data. LYL, DDW, and ZCM gave invaluable advices in design, experiment performing, and paper writing. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by Yangtze Normal University. Participants provided signed informed consent. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Communicated by Yuan Ge.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liao, J., Luo, L., Zhang, L. et al. Comparison of the effects of three fungicides on clubroot disease of tumorous stem mustard and soil bacterial community. J Soils Sediments 22, 256–271 (2022). https://doi.org/10.1007/s11368-021-03073-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-021-03073-z