Abstract
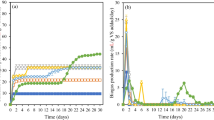
A blend of organic municipal solid waste, slaughterhouse waste, fecal sludge, and landfill leachate was selected in different mixing ratios to formulate the best substrate mixture for biomethanation. Individual substrates were characterized, and the mixing ratio was optimized with the help of a response surface methodology tool to a value of 1:1:1:1 (with a C/N ratio of 28±0.769 and total volatile fatty acid (VFA) concentration of 2500±10.53 mg/L) to improve the overall biomethanation. The optimized blend (C/N ratio: 28.6, VFA: 2538 mg/L) was characterized for physicochemical, biological, and microbial properties and subjected to anaerobic digestion in lab-scale reactors of 1000 mL capacity with and without the addition of inoculum. The biogas yield of individual substrates and blends was ascertained separately. The observed cumulative biogas yield over 21 days from the non-inoculated substrates varied between 142±1.95 mL (24.6±0.3 ml/gVS) and 1974.5±21.72 mL (270.4±3.1 ml/gVS). In comparison, the addition of external inoculation at a 5% rate (w/w) of the substrate uplifted the minimum and maximum cumulative gas yield values to 203±9.9 mL (35.0±1.6 mL/gVS) and 3394±13.4 mL (315.3±1.2 mL/gVS), respectively. The inoculum procured from the Defence Research and Development Organisation (DRDO) was screened in advance, considering factors such as maximizing VFA production and consumption rate, biogas yield, and digestate quality. A similar outcome regarding biogas yield and digestate quality was observed for the equivalent blend. The cumulative gas yield increased from 2673±14.5 mL (373.7±2.2 mL/gVS) to 4284±111.02 mL (391.47±20.02 mL/gVS) over 21 days post-application of a similar dosage of DRDO inoculum. The 16S rRNA genomic analysis revealed that the predominant bacterial population belonged to the phylum Firmicutes, with the majority falling within the orders Clostridiales and Lactobacillales. Ultimately, the study advocates the potential of the blend mentioned above for biomethanation and concomitant enrichment of both biogas yield and digestate quality.










Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Adedara ML, Taiwo R, Bork HR (2023) Municipal solid waste collection and coverage rates in sub-Saharan African countries: a comprehensive systematic review and meta-analysis. Waste 1(2):389–413. https://doi.org/10.3390/waste1020024
Ahmed B, Gahlot P, Balasundaram G, Tyagi VK, Vivekanand V, Kazmi AA (2023) Semi-continuous anaerobic co-digestion of thermal and thermal-alkali processed organic fraction of municipal solid waste: methane yield, energy analysis, anaerobic microbiome. J Environ Manag 345:118907. https://doi.org/10.1016/j.jenvman.2023.118907
Alavi-Borazjani SA, Capela I, Tarelho LA (2020) Over-acidification control strategies for enhanced biogas production from anaerobic digestion: a review. Biomass Bioenergy 143:105833. https://doi.org/10.1016/j.biombioe.2020.105833
Al-Sulaimi IN, Nayak JK, Alhimali H, Sana A, Al-Mamun A (2022) Effect of volatile fatty acids accumulation on biogas production by sludge-feeding thermophilic anaerobic digester and predicting process parameters. Fermentation 8(4):184. https://doi.org/10.3390/fermentation8040184
Amelia TSM, Lau NS, Amirul AAA, Bhubalan K (2020) Metagenomic data on bacterial diversity profiling of high-microbial-abundance tropical marine sponges Aaptos aaptos and Xestospongia muta from waters off Terengganu, South China Sea. Data Brief 31:105971. https://doi.org/10.1016/j.dib.2020.105971
Andriamanohiarisoamanana FJ, Saikawa A, Kan T, Qi G, Pan Z, Yamashiro T, Iwasaki M, Ihara I, Nishida T, Umetsu K (2018) Semi-continuous anaerobic co-digestion of dairy manure, meat and bone meal and crude glycerol: Process performance and digestate valorization. Renew Energy 128:1–8. https://doi.org/10.1016/j.renene.2018.05.056
Awasthi MK, Sarsaiya S, Wainaina S, Rajendran K, Awasthi SK, Liu T, Duan Y, Jain A, Sindhu R, Binod P, Pandey A, Zhang Z, Taherzadeh MJ (2021) Techno-economics and life-cycle assessment of biological and thermochemical treatment of bio-waste. Renew Sust Energ Rev 144:110837. https://doi.org/10.1016/j.rser.2021.110837
Azevedo A, Lapa N, Moldão M, Duarte E (2023) Opportunities and challenges in the anaerobic co-digestion of municipal sewage sludge and fruit and vegetable wastes: a review. Energy Nexus 10:100202. https://doi.org/10.1016/j.nexus.2023.100202
Bhatia L, Jha H, Sarkar T, Sarangi PK (2023) Food waste utilization for reducing carbon footprints towards sustainable and cleaner environment: a review. Int J Environ Res Public Health 20(3):2318. https://doi.org/10.3390/ijerph20032318
Capodaglio AG, Callegari A (2023) Energy and resources recovery from excess sewage sludge: a holistic analysis of opportunities and strategies. Resour Conserv Recycl Adv 19:200184. https://doi.org/10.1016/j.rcradv.2023.200184
Choi Y, Ryu J, Lee SR (2020) Influence of carbon type and carbon to nitrogen ratio on the biochemical methane potential, pH, and ammonia nitrogen in anaerobic digestion. J Anim Sci Technol 62(1):74. https://doi.org/10.5187/jast.2020.62.1.74
Choudhury AR, Boyina LP, Kumar DL, Singh N, Palani SG, Mehdizadeh M, Kumar MVP, Leelavathi A, Rao BK, Begum SUA, Patrick K (2022) Biomined and fresh municipal solid waste as sources of refuse derived fuel. In: Pathak P, Palani SG (eds) Circular economy in municipal solid waste landfilling: biomining & leachate treatment. Radionuclides and Heavy Metals in the Environment. Springer, Cham. https://doi.org/10.1007/978-3-031-07785-2_11
Choudhury AR, Kumar NA, Arutchelvan V, Srinivas K (2018b) Recovery of agro-based bio-products from cattle farm waste using aerobic fluidized crystallization reactor. Biotechnol Res 4(4):119–126
Choudhury AR, Natarajan AK, Kesavarapu S, Veeraraghavan A, Dugyala SK, Rao K, Thota KR (2018a) Technical feasibility of Hermetia illucens in integrated waste management, renovated with sewage water, an overview. Open Access Libr J 5(04):e4421. https://doi.org/10.4236/oalib.1104421
Choudhury AR, Singh N, Palani SG, Lalwani J (2023) Bioremediation of reverse osmosis concentrate generated from the treatment of landfill leachate. Environ Sci Pollut Res 30(41):93934–93951. https://doi.org/10.1007/s11356-023-28957-0
Clarke WP, Xie S, Patel M (2016) Rapid digestion of shredded MSW by sequentially flooding and draining small landfill cells. Waste Manag 55:12–21. https://doi.org/10.1016/j.wasman.2015.11.050
Czekała W, Nowak M, Piechota G (2023) Sustainable management and recycling of anaerobic digestate solid fraction by composting: a review. Bioresour Technol 375:128813. https://doi.org/10.1016/j.biortech.2023.128813
da Silva GH, Barros NO, Santana LA, Carneiro JD, Otenio MH (2021) Shifts of acidogenic bacterial group and biogas production by adding two industrial residues in anaerobic co-digestion with cattle manure. J Environ Sci Health - Toxic/Hazard Subst Environ Eng 56:1503–1511. https://doi.org/10.1080/10934529.2021.2015987
da Silva Mazareli RC, Duda RM, Leite VD, de Oliveira RA (2016) Anaerobic co-digestion of vegetable waste and swine wastewater in high-rate horizontal reactors with fixed bed. Waste Manag 52:112–121. https://doi.org/10.1016/j.wasman.2016.03.021
Defence Research and Development Organisation (DRDO) (2019) BIO-DIGESTER. Retrieved September 12, 2023, from https://www.drdo.gov.in/bio-digester
Demichelis F, Tommasi T, Deorsola FA, Marchisio D, Fino D (2022) Effect of inoculum origin and substrate-inoculum ratio to enhance the anaerobic digestion of organic fraction municipal solid waste (OFMSW). J Clean Prod 351:131539. https://doi.org/10.1016/j.jclepro.2022.131539
Fikri Hamzah MA, Abdul PM, Mahmod SS, Azahar AM, Jahim JM (2020) Performance of anaerobic digestion of acidified palm oil mill effluent under various organic loading rates and temperatures. Water 12(9):2432. https://doi.org/10.3390/w12092432
Filer J, Ding HH, Chang S (2019) Biochemical methane potential (BMP) assay method for anaerobic digestion research. Water 11(5):921. https://doi.org/10.3390/w11050921
Garcia-Peña EI, Parameswaran P, Kang DW, Canul-Chan M, Krajmalnik-Brown R (2011) Anaerobic digestion and co-digestion processes of vegetable and fruit residues: process and microbial ecology. Bioresour Technol 102(20):9447–9455. https://doi.org/10.1016/j.biortech.2011.07.068
Gates RS, Batista MD, Souza CF, Tinoco IF, Miranda S, Álvares FF (2014) Using cultures of “Effective Microorganisms” (EM) as inoculum in substrates for anaerobic digestion. In 2014 Montreal, Quebec Canada, p. 1. American Society of Agricultural and Biological Engineers. July 13–July 16, 2014, https://doi.org/10.13031/aim.20141913519
Griffin ME, McMahon KD, Mackie RI, Raskin L (1998) Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol Bioeng 57(3):342–355. https://doi.org/10.1002/(SICI)1097-0290(19980205)57:3%3C342::AID-BIT11%3E3.0.CO;2-I
Ijoma GN, Nkuna R, Mutungwazi A, Rashama C, Matambo TS (2021) Applying PICRUSt and 16S rRNA functional characterisation to predicting co-digestion strategies of various animal manures for biogas production. Sci Rep 11(1):19913. https://doi.org/10.1038/s41598-021-99389-4
Johnravindar D, Wong JW, Chakraborty D, Bodedla G, Kaur G (2021) Food waste and sewage sludge co-digestion amended with different biochars: VFA kinetics, methane yield and digestate quality assessment. J Environ Manag 290:112457. https://doi.org/10.1016/j.jenvman.2021.112457
Joshi R, Ahmed S (2016) Status and challenges of municipal solid waste management in India: a review. Cogent Environ Sci 2(1):1139434. https://doi.org/10.1080/23311843.2016.1139434
Karak T, Bhagat RM, Bhattacharyya P (2012) Municipal solid waste generation, composition, and management: the world scenario. Crit Rev Environ Sci Technol 42(15):1509–1630. https://doi.org/10.1080/10643389.2011.569871
Khadka A, Parajuli A, Dangol S, Thapa B, Sapkota L, Carmona-Martínez AA, Ghimire A (2022) Effect of the substrate to inoculum ratios on the kinetics of biogas production during the mesophilic anaerobic digestion of food waste. Energies 15(3):834. https://doi.org/10.3390/en15030834
Kim BR, Shin J, Guevarra RB, Lee JH, Kim DW, Seol KH, Lee JH, Kim HB, Isaacson RE (2017) Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol 27(12):2089–2093. https://doi.org/10.4014/jmb.1709.09027
Koch K, Hafner SD, Astals S, Weinrich S (2020) Evaluation of common supermarket products as positive controls in biochemical methane potential (BMP) tests. Water 12(5):1223. https://doi.org/10.3390/w12051223
Kumar A, Agrawal A (2020) Recent trends in solid waste management status, challenges, and potential for the future Indian cities–a review. Curr Res Environ Sustain 2:100011. https://doi.org/10.1016/j.crsust.2020.100011
Li Y, Zhao J, Krooneman J, Euverink GJW (2021) Strategies to boost anaerobic digestion performance of cow manure: Laboratory achievements and their full-scale application potential. Sci Total Environ 755:142940. https://doi.org/10.1016/j.scitotenv.2020.142940
Loganath R, Senophiyah-Mary J (2020) Critical review on the necessity of bioelectricity generation from slaughterhouse industry waste and wastewater using different anaerobic digestion reactors. Renew Sust Energ Rev 134:110360. https://doi.org/10.1016/j.rser.2020.110360
Maurus K, Kremmeter N, Ahmed S, Kazda M (2023) High-resolution monitoring of VFA dynamics reveals process failure and exponential decrease of biogas production. Biomass Convers Biorefin 13:10653–10663. https://doi.org/10.1007/s13399-021-02043-2
Mo R, Guo W, Batstone D, Makinia J, Li Y (2023) Modifications to the anaerobic digestion model no. 1 (ADM1) for enhanced understanding and application of the anaerobic treatment processes–a comprehensive review. Water Res 244:120504. https://doi.org/10.1016/j.watres.2023.120504
Mozhiarasi V, Natarajan TS (2022) Slaughterhouse and poultry wastes: management practices, feedstocks for renewable energy production, and recovery of value added products. Biomass Convers Biorefin 1-24. https://doi.org/10.1007/s13399-022-02352-0
Mukherjee S, Mukhopadhyay S, Hashim MA, Sen Gupta B (2015) Contemporary environmental issues of landfill leachate: assessment and remedies. Crit Rev Environ Sci Technol 45(5):472–590. https://doi.org/10.1080/10643389.2013.876524
Mutungwazi A, Ijoma GN, Matambo TS (2021) The significance of microbial community functions and symbiosis in enhancing methane production during anaerobic digestion: a review. Symbiosis 83:1–24. https://doi.org/10.1007/s13199-020-00734-4
Neshat SA, Mohammadi M, Najafpour GD, Lahijani P (2017) Anaerobic co-digestion of animal manures and lignocellulosic residues as a potent approach for sustainable biogas production. Renew Sust Energ Rev 79:308–322. https://doi.org/10.1016/j.rser.2017.05.137
Ortiz-Burgos S (2016) Shannon-Weaver diversity index. In: Kennish MJ (ed) Encyclopedia of estuaries. Encyclopedia of earth sciences series. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-8801-4_233
Pan X, Zhao L, Li C, Angelidaki I, Lv N, Ning Cai G, Zhu G (2021) Deep insights into the network of acetate metabolism in anaerobic digestion: focusing on syntrophic acetate oxidation and homoacetogenesis. Water Res 190:116774. https://doi.org/10.1016/j.watres.2020.116774
Pirsaheb M, Hossaini H, Amini J (2019) Evaluation of a zeolite/anaerobic buffled reactor hybrid system for treatment of low bio-degradable effluents. Mater Sci Eng C 104:109943. https://doi.org/10.1016/j.msec.2019.109943
Qin S, Wainaina S, Liu H, Soufiani AM, Pandey A, Zhang Z, Awasthi MK, Taherzadeh MJ (2021) Microbial dynamics during anaerobic digestion of sewage sludge combined with food waste at high organic loading rates in immersed membrane bioreactors. Fuel 303:121276. https://doi.org/10.1016/j.fuel.2021.121276
Qiu L, Deng YF, Wang F, Davaritouchaee M, Yao YQ (2019) A review on biochar-mediated anaerobic digestion with enhanced methane recovery. Renew Sust Energ Rev 115:109373. https://doi.org/10.1016/j.rser.2019.109373
Sarker S, Lamb JJ, Hjelme DR, Lien KM (2019) A review of the role of critical parameters in the design and operation of biogas production plants. Appl Sci 9(9):1915. https://doi.org/10.3390/app9091915
Selormey GK, Barnes B, Kemausuor F, Darkwah L (2021) A review of anaerobic digestion of slaughterhouse waste: effect of selected operational and environmental parameters on anaerobic biodegradability. Rev Environ Sci Biotechnol 20:1073–1086. https://doi.org/10.1007/s11157-021-09596-8
Shahab S, Anjum M (2022) Solid waste management scenario in India and illegal dump detection using deep learning: an AI approach towards the sustainable waste management. Sustainability 14(23):15896. https://doi.org/10.3390/su142315896
Sharma HB, Vanapalli KR, Samal B, Cheela VRS, Dubey BK, Bhattacharya J (2021) Circular economy approach in solid waste management system to achieve UN-SDGs: Solutions for post-COVID recovery. Sci Total Environ 800:149605. https://doi.org/10.1016/j.scitotenv.2021.149605
Sheets JP, Yang L, Ge X, Wang Z, Li Y (2015) Beyond land application: emerging technologies for the treatment and reuse of anaerobically digested agricultural and food waste. Waste Manag 44:94–115. https://doi.org/10.1016/j.wasman.2015.07.037
Shet R, Mutnuri S (2021) Anaerobic co-digestion of organic fraction of municipal solid waste and septage for sustainable waste treatment: a case study from Goa, India. Eur J Energy Res 1(3):1–8. https://doi.org/10.24018/ejenergy.2021.1.3.15
Sicchieri IM, de Quadros TCF, Bortoloti MA, Fernandes F, Kuroda EK (2022) Selection, composition, and validation of standard inoculum for anaerobic digestion assays. Biomass Bioenergy 164:106558. https://doi.org/10.1016/j.biombioe.2022.106558
Silvestre G, Fernández B, Bonmatí A (2015) Addition of crude glycerine as strategy to balance the C/N ratio on sewage sludge thermophilic and mesophilic anaerobic co-digestion. Bioresour Technol 193:377–385. https://doi.org/10.1016/j.biortech.2015.06.098
Singh D, Tembhare M, Machhirake N, Kumar S (2023) Impact of municipal solid waste landfill leachate on biogas production rate. J Environ Manag 336:117643. https://doi.org/10.1016/j.jenvman.2023.117643
Srivastava AN, Chakma S (2023) Lime sludge assisted anaerobic bioreactor landfilling of municipal solid waste for enhanced leachate stabilization and bioenergy yield. Process Saf Environ Prot 178:278–286. https://doi.org/10.1016/j.psep.2023.08.039
Velvizhi G, Shanthakumar S, Das B, Pugazhendhi A, Priya TS, Ashok B, Nanthagopal K, Vignesh R, Karthick C (2020) Biodegradable and non-biodegradable fraction of municipal solid waste for multifaceted applications through a closed loop integrated refinery platform: paving a path towards circular economy. Sci Total Environ 731:138049. https://doi.org/10.1016/j.scitotenv.2020.138049
Wang R, Gmoser R, Taherzadeh MJ, Lennartsson PR (2021) Solid-state fermentation of stale bread by an edible fungus in a semi-continuous plug-flow bioreactor. Biochem Eng J 169:107959. https://doi.org/10.1016/j.bej.2021.107959
Wang Z, Hu Y, Wang S, Wu G, Zhan X (2023) A critical review on dry anaerobic digestion of organic waste: characteristics, operational conditions, and improvement strategies. Renew Sust Energ Rev 176:113208. https://doi.org/10.1016/j.rser.2023.113208
Weldon S, Rivier PA, Joner EJ, Coutris C, Budai A (2022) Co-composting of digestate and garden waste with biochar: effect on greenhouse gas production and fertilizer value of the matured compost. Environ Technol 44(28):4261–4271. https://doi.org/10.1080/09593330.2022.2089057
Xiong H, Chen J, Wang H, Shi H (2012) Influences of volatile solid concentration, temperature and solid retention time for the hydrolysis of waste activated sludge to recover volatile fatty acids. Bioresour Technol 119:285–292. https://doi.org/10.1016/j.biortech.2012.05.126
Yadav M, Joshi C, Paritosh K, Thakur J, Pareek N, Masakapalli SK, Vivekanand V (2022) Reprint of Organic waste conversion through anaerobic digestion: a critical insight into the metabolic pathways and microbial interactions. Metab Eng 71:62–76. https://doi.org/10.1016/j.ymben.2022.02.001
Yan Z, Song Z, Li D, Yuan Y, Liu X, Zheng T (2015) The effects of initial substrate concentration, C/N ratio, and temperature on solid-state anaerobic digestion from composting rice straw. Bioresour Technol 177:266–273. https://doi.org/10.1016/j.biortech.2014.11.089
Zamri MFMA, Hasmady S, Akhiar A, Ideris F, Shamsuddin AH, Mofijur M, Fattah IMR, Mahlia TMI (2021) A comprehensive review on anaerobic digestion of organic fraction of municipal solid waste renew. Sust Energ Rev 137:110637. https://doi.org/10.1016/j.rser.2020.110637
Zhang Y, Li C, Yuan Z, Wang R, Angelidaki I, Zhu G (2023) Syntrophy mechanism, microbial population, and process optimization for volatile fatty acids metabolism in anaerobic digestion. Chem Eng J 452:139137. https://doi.org/10.1016/j.cej.2022.139137
Zhang Z, Liu R, Lan Y, Zheng W, Chen L (2024) Anaerobic co-fermentation of waste activated sludge with corn gluten meal enhanced phosphorus release and volatile fatty acids production: critical role of corn gluten meal dosage on fermentation stages and microbial community traits. Bioresour Technol 394:130275. https://doi.org/10.1016/j.biortech.2023.130275
Acknowledgements
The present research was supported by Banka Bio Limited. The authors are grateful to the organization for extending access to their FSTP and laboratory facilities.
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Atun Roy Choudhury. The first draft of the manuscript was written by Atun Roy Choudhury, Neha Singh, Sankar Ganesh Palani, Hemapriya Srinivasan, and Jitesh Lalwani, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Ta Yeong Wu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choudhury, A.R., Singh, N., Lalwani, J. et al. Enhancing biomethanation performance through co-digestion of diverse organic wastes: a comprehensive study on substrate optimization, inoculum selection, and microbial community analysis. Environ Sci Pollut Res (2024). https://doi.org/10.1007/s11356-024-33557-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11356-024-33557-7