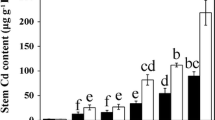
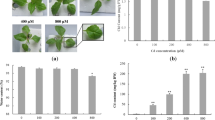
Abstract
Siegesbeckia orientalis L. was identified as a novel Cd-hyperaccumulator and valuable phytoremediation material. However, the molecular mechanisms underlying Cd accumulation in S. orientalis are largely unknown. In this study, RNA-Seq analysis was performed to study the Cd-accumulating mechanisms in its roots with or without Cd treatment. The RNA-seq analysis generated 312 million pairs of clean reads and 78G sequencing data. De novo transcriptome assembly produced 355,070 transcripts with an average length of 823.59 bp and 194,207 unigenes with an average length of 605.68 bp. Comparative transcriptome analyses identified a large number of differentially expressed genes in roots under Cd stress, and functional annotation suggested that S. orientalis utilizes various biological pathways involving many gene networks working simultaneously to cope with the stress. This study revealed that four biological pathways were mainly involved in S. orientalis tolerance to Cd stress, including reactive oxygen species scavenging, phenylpropanoid biosynthesis pathway, Cd absorption and transport, and ABA signaling pathway. The genes related to photosynthesis and heavy metal transport are likely the potential candidates and could be further investigated to determine their roles in Cd tolerance in S. orientalis roots. These findings will be useful to understand the Cd accumulation mechanisms in S. orientalis and facilitate the study of phytoremediation at the molecular level in plants.







Similar content being viewed by others
References
Agrawal SB, Mishra S (2009) Effects of supplemental ultraviolet-B and cadmium on growth, antioxidants and yield of Pisum sativum L. Ecotoxicol Environ Saf 72:610–618
Ali H, Khan E, Sajad MA (2013) Phytoremediation of heavy metals-concepts and applications. Chemosphere 91:869–881
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Cao F, Chen F, Sun H, Zhang G, Chen ZH, Wu F (2014) Genome-wide transcriptome and functional analysis of two contrasting genotypes reveals key genes for cadmium tolerance in barley. BMC Genomics 155:611
Chen J, Yang L, Yan X, Liu Y, Wang R, Fan T, Ren Y, Tang X, Xiao F, Liu Y, Cao S (2016) Zinc-finger transcription factor ZAT6 positively regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis. Plant Physiol 171:707–719
Clemens S (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88:1707–1719
Farinati S, DalCorso G, Varotto S, Furini A (2010) The Brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants. New Phytol 185:964–978
Feng J, Jia W, Lv S, Bao H, Miao F, Zhang X, Wang J, Li J, Li D, Zhu C, Li S, Li Y (2018) Comparative transcriptome combined with morpho-physiological analyses revealed key factors for differential cadmium accumulation in two contrasting sweet sorghum genotypes. Plant Biotechnol J 16:558–571
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37
Fusco N, Micheletto L, Dal Corso G, Borgato L, Furini A (2005) Identification of cadmium-regulated genes by cDNA-AFLP in the heavy metal accumulator Brassica juncea L. J Exp Bot 56:3017–3027
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
He F, Liu Q, Zheng L, Cui Y, Shen Z, Zheng L (2015) RNA-Seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front Plant Sci 6:1136
Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette ML, Cuine S, Auroy P, Richaud P, Forestier C, Bourguignon J, Renou JP, Vavasseur A, Leonhardt N (2006) Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88:1751–1765
Hou D, O'Connor D, Nathanail P, Tian L, Ma Y (2017) Integrated GIS and multivariate statistical analysis for regional scale assessment of heavy metal soil contamination: a critical review. Environ Pollut 231:1188–1200
Ismael MA, Elyamine AM, Moussa MG, Cai M, Zhao X, Hu C (2019) Cadmium in plants: uptake, toxicity, and its interactions with selenium fertilizers. Metallomics 11:255–277
Jansson S (1994) The light-harvesting chlorophyll a/b-binding proteins. Biochim Biophys Acta 1184:1–19
Jean ML, Schikora A, Mari S, Briat JF, Curie C (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44:769–782
Khan A, Khan S, Alam M, Khan MA, Aamir M, Qamar Z, Ur Rehman Z, Perveen S (2016) Toxic metal interactions affect the bioaccumulation and dietary intake of macro- and micro-nutrients. Chemosphere 146:121–128
Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50:207–218
Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52:2136–2146
Koen E, Besson-Bard A, Duc C, Astier J, Gravot A, Richaud P, Lamotte O, Boucherez J, Gaymard F, Wendehenne D (2013) Arabidopsis thaliana nicotianamine synthase 4 is required for proper response to iron deficiency and to cadmium exposure. Plant Sci 209:1–11
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345–W349
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Li A, Zhang J, Zhou Z (2014) PLEK: a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 15:311
Li C, Yang X, Xu Y, Li L, Wang Y (2018) Cadmium detoxification induced by salt stress improves cadmium tolerance of multi-stress-tolerant Pichia kudriavzevii. Environ Pollut 242:845–854
Liu L, Wei J, Ruan J (2017) Pathway enrichment analysis with networks. Genes (Basel) 8
Martin F, Bovet L, Cordier A, Stanke M, Gunduz I, Peitsch MC, Ivanov NV (2012) Design of a tobacco exon array with application to investigate the differential cadmium accumulation property in two tobacco varieties. BMC Genomics 13:674
Ridley BL, O'Neill MA, Mohnen D (2001) Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929–967
Rizwan M, Ali S, Adrees M, Ibrahim M, Tsang DCW, Zia-Ur-Rehman M, Zahir ZA, Rinklebe J, Tack FMG, Ok YS (2017) A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 182:90–105
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Romero-Puertas MC, Corpas FJ, Rodriguez-Serrano M, Gomez M, Del Rio LA, Sandalio LM (2007) Differential expression and regulation of antioxidative enzymes by cadmium in pea plants. J Plant Physiol 164:1346–1357
Shamsi IH, Zhang GP, Hu HL, Xue QY, Hussain N, Ali E, Shen QF, Zheng WT, Zhang QC, Liu XX (2014) Assessment of the hazardous effects of Cd on physiological and biochemical characteristics of soybean genotypes. Int J Agric Biol 29:77–79
Song X, Hu X, Ji P, Li Y, Chi G, Song Y (2012) Phytoremediation of cadmium-contaminated farmland soil by the hyperaccumulator Beta vulgaris L. var. cicla. Bull Environ Contam Toxicol 88:623–626
Song B, Zeng G, Gong J, Liang J, Xu P, Liu Z, Zhang Y, Zhang C, Cheng M, Liu Y, Ye S, Yi H, Ren X (2017) Evaluation methods for assessing effectiveness of in situ remediation of soil and sediment contaminated with organic pollutants and heavy metals. Environ Int 105:43–55
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41:e166
Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK (2011) The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot 62:4843–4850
Tamas L, Dudikova J, Durcekova K, Haluskova L, Huttova J, Mistrik I, Olle M (2008) Alterations of the gene expression, lipid peroxidation, proline and thiol content along the barley root exposed to cadmium. J Plant Physiol 165:1193–1203
Toth G, Hermann T, Da Silva MR, Montanarella L (2016) Heavy metals in agricultural soils of the European Union with implications for food safety. Environ Int 88:299–309
Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6:rs8
Wang Z, Zhang Y, Huang Z, Lin H (2008) Antioxidative response of metal-accumulator and non-accumulator plants under cadmium stress. Plant Soil 310:137–149
Wang P, Deng X, Huang Y, Fang X, Zhang J, Wan H, Yang C (2015) Comparison of subcellular distribution and chemical forms of cadmium among four soybean cultivars at young seedlings. Environ Sci Pollut Res Int 22:19584–19595
Weber M, Trampczynska A, Clemens S (2006) Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd(2+)-hypertolerant facultative metallophyte Arabidopsis halleri. Plant Cell Environ 29:950–963
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322
Xu X-X, Dong YY, Deng YL, Huo QL, Zhang SR, Yu-Lin PU (2016) Effects of cadmium stress on growth and photosynthetic parameters of Sigesbeckia orientalis L. J Agro-Environ Sci
Xu ZM, Li QS, Yang P, Ye HJ, Chen ZS, Guo SH, Wang LL, He BY, Zeng EY (2017) Impact of osmoregulation on the differences in cd accumulation between two contrasting edible amaranth cultivars grown on cd-polluted saline soils. Environ Pollut 224:89–97
Xu X, Zhang S, Xian J, Yang Z, Cheng Z, Li T (2018) Effects of cadmium stress on growth and photosynthetic parameters of and detoxification in Siegesbeckia orientalis L. Int J Phytoremediation 20:973–980
Yan H et al (2019) Author correction: Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat Commun 10:3301
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14
Yousaf B, Liu G, Wang R, Imtiaz M, Zia-Ur-Rehman M, Munir MA, Niu Z (2016) Bioavailability evaluation, uptake of heavy metals and potential health risks via dietary exposure in urban-industrial areas. Environ Sci Pollut Res Int 23:22443–22453
Yue R, Lu C, Qi J, Han X, Yan S, Guo S, Liu L, Fu X, Chen N, Yin H, Chi H, Tie S (2016) Transcriptome analysis of cadmium-treated roots in maize (Zea mays L.). Front Plant Sci 7:1298
Zhang M, Liu X, Yuan L, Wu K, Duan J, Wang X, Yang L (2012) Transcriptional profiling in cadmium-treated rice seedling roots using suppressive subtractive hybridization. Plant Physiol Biochem 50:79–86
Zhang S, Lin H, Deng L, Gong G, Jia Y, Xiaoxun XU, Ting LI, Yun LI, Hui C (2013) Cadmium tolerance and accumulation characteristics of Siegesbeckia orientalis L. Ecol Eng 51:133–139
Zhou Q, Guo JJ, He CT, Shen C, Huang YY, Chen JX, Guo J, Yuan JG, Yang ZY (2016) Comparative transcriptome analysis between low- and high-cadmium-accumulating genotypes of pakchoi (Brassica chinensis L.) in response to cadmium stress. Environ Sci Technol 50:6485
Acknowledgments
This work was supported by The Key Research and Development Projects of Sichuan Province, China (No. 2019YFN0020) and The Science and Technology Project for Sichuan Environmental Protection (No. 2018HB30). We thank Zhien Pu, Zhongxu Chen, Peng Liu, Yulan Deng, and Yuanyuan Dong of Sichuan Agricultural University for supporting the transcriptome analysis and assisting with the research.
Author information
Authors and Affiliations
Contributions
XX and SZ conceived the study and designed the experiments. SZ supervised the research, acquired funding, and revised the manuscript. XX, ZC, TL, YJ, and GW performed the experiments and analyzed the data; XX, ZY, JX, YY, and WZ cultivated and prepared the plant materials; and XX wrote the manuscript. All of the authors read and approved the final manuscript.
Corresponding author
Additional information
Responsible editor: Gangrong Shi
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(XLSX 3.34 MB)
Rights and permissions
About this article
Cite this article
Xu, X., Zhang, S., Cheng, Z. et al. Transcriptome analysis revealed cadmium accumulation mechanisms in hyperaccumulator Siegesbeckia orientalis L.. Environ Sci Pollut Res 27, 18853–18865 (2020). https://doi.org/10.1007/s11356-020-08387-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-08387-y