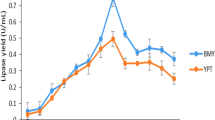
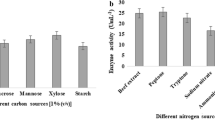
Abstract
Lipase, a versatile hydrolytic enzyme, is gaining more importance in environmental applications such as treatment of oil and grease containing wastewater, pretreatment of solid waste/industrial wastewater for anaerobic treatment. In the present study, the attempts have been made to improve the production of lipase from Staphylococcus hominis MTCC 8980 by optimization of pH, temperature, and agitation speed in lab scale shake flasks culture. The experiments were designed using the full factorial central composite design of experiment. A total of 20 experiments were conducted, and the optimized pH, temperature, and agitation speed were found to be 7.9, 33.1 °C, and 178.4 rpm, respectively. The results of the analysis of variance (ANOVA) test revealed that the linear terms for temperature and agitation were significant (p value < 0.05). Interaction for pH and agitation speed was found to have a significant effect on lipase production from S. hominis MTCC 8980. A 150% increase in enzyme activity was observed under the optimized conditions with the maximum lipase activity of 1.82 U/ml. Further enhancement of enzyme activity can be expected from the optimization of medium components.




Similar content being viewed by others
References
Basheer SM, Chellappan S, Beena PS, Sukumaran RK, Elyas KK, Chandrasekaran M (2011) Lipase from marine Aspergillus awamori BTMFW032 production, partial purification and application in oil effluent treatment. New Biotechnol 28(6):627–638
Bharathi D, Rajalakshmi G, Komathi S (2018) Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil. J King Saud Univ-Sci
Bosley JA, Clayton JC (1994) Blueprint for a lipase support use of hydrophobic controlled pore glasses as model systems. Biotechnol Bioeng 43:934–938
Charton E, Macrae AR (1992) Substrate specificities for lipases A and B from Geotrichum candidum CMICC 33546. Biochim Biophys Acta 1123:59–64
Chauhan M, Chauhan RS, Garlapati VK (2013) Modeling and optimization studies on a novel lipase production by Staphylococcus arlettae through submerged fermentation. Enzyme Res 2013:1–8
Colla LM, Primaz AL, Benedetti S, Loss RA, Lima MD, Reinehr CO, Bertolin TE, Costa JA (2016) Surface response methodology for the optimization of lipase production under submerged fermentation by filamentous fungi. Braz J Microbiol 47(2):461–467
Dharmsthiti S, Kuhasuntisuk B (1998) Lipase from Pseudomonas aeruginosa LP602 biochemical properties and application for wastewater treatment. J Ind Microbiol Biotechnol 21(1–2):75–80
Dutta K, Dasu VV, Hegde K (2013) Development of medium and kinetic modeling for enhanced production of cutinase from Pseudomonas cepacia NRRL B-2320. Adv Microbiol 3(6):479. https://www.marketsandmarkets.com/PressReleases/lipase.asp
Javed S, Azeem F, Hussain S, Rasul I, Siddique MH, Riaz M, Nadeem H (2018) Bacterial lipases: a review on purification and characterization. Prog Biophys Mol Biol 132:23–34
Kanmani P, Aravind J, Kumaresan K (2015) An insight into microbial lipases and their environmental facet. Int J Appl Biol Pharm 2(3):1147–1162
Kaushik R, Saran S, Isar J, Saxena RK (2006) Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus. J Mol Catal B Enzym 40(3–4):121–126
Larbidaouadi K, Benattouche Z, Abbouni B (2015) Screening selection identification production and optimization of bacterial lipase isolated from industrial rejection of gas station. Int J Biotechnol Allied Fields 3(9):146–153
Liu CH, Lu WB, Chang JS (2006) Optimizing lipase production of Burkholderia sp. by response surface methodology. Process Biochem 41(9):1940–1944
Mahdi BA, Bhattacharya A, Gupta A (2012) Enhanced lipase production from Aeromonas sp. S1 using Sal deoiled seed cake as novel natural substrate for potential application in dairy wastewater treatment. J Chem Technol Biotechnol 87(3):418–426
Marimuthu K (2013) Isolation and characterization of Staphylococcus hominis JX961712 from oil contaminated soil. Int J Pharm Sci Res 7(3):252–256
Momsia T, Momsia P (2013) A review on microbial lipase-versatile tool for industrial applications. Int J life Sci Biotechnol Pharma Res 2(4):2250–3137
Moon HC, Song IS (2011) Enzymatic hydrolysis of foodwaste and methane production using UASB bioreactor. Int J Green Energy 8(3):361–371
de Morais WG Jr, Kamimura ES, Ribeiro EJ, Pessela BC, Cardoso VL, de Resende MM (2016) Optimization of the production and characterization of lipase from Candida rugosa and Geotrichum candidum in soybean molasses by submerged fermentation. Protein Expr Purif 123:26–34
Pignède G, Wang H, Fudalej F, Gaillardin C, Seman M, Nicaud JM (2000) Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol 182(10):2802–2810
Prasad MP, Manjunath K (2011) Comparative study on biodegradation of lipid-rich wastewater using lipase producing bacterial species. Indian J Biotechnol 10(1):121–124
Sharma A, Bardhan D, Patel R (2009) Optimization of physical parameters for lipase production from Arthrobacter sp. BGCC# 490. Indian J Biochem Biophys 46(2):178–183
Singh R, Kumar M, Mittal A, Mehta PK (2016) Microbial enzymes: industrial progress in 21st century 3. Biotech 6(2):174
Teng Y, Xu Y (2008) Culture condition improvement for whole-cell lipase production in submerged fermentation by Rhizopus chinensis using statistical method. Bioresour Technol 99(9):3900–3907
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We have followed the accepted principles of ethical and professional conduct.
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights and informed consent
The work did not involve human participants and/or animals.
Additional information
Responsible editor: Philippe Garrigues
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Behera, A.R., Veluppal, A. & Dutta, K. Optimization of physical parameters for enhanced production of lipase from Staphylococcus hominis using response surface methodology. Environ Sci Pollut Res 26, 34277–34284 (2019). https://doi.org/10.1007/s11356-019-04304-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-04304-0