Abstract
Introduction
Previous studies have explored prediction value of serum metabolites in neoadjuvant chemoradiation therapy (NCRT) response for rectal cancer. To date, limited literature is available for serum metabolome changes dynamically through NCRT.
Objectives
This study aimed to explore temporal change pattern of serum metabolites during NCRT, and potential metabolic biomarkers to predict the pathological response to NCRT in locally advanced rectal cancer (LARC) patients.
Methods
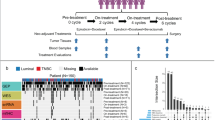
Based on dynamic UHPLC-QTOF-MS untargeted metabolomics design, this study included 106 LARC patients treated with NCRT. Biological samples of the enrolled patients were collected in five consecutive time-points. Untargeted metabolomics was used to profile serum metabolic signatures from LARC patients. Then, we used fuzzy C-means clustering (FCM) to explore temporal change patterns in metabolites cluster and identify monotonously changing metabolites during NCRT. Repeated measure analysis of variance (RM-ANOVA) and multilevel partial least-squares discriminant analysis (ML-PLS-DA) were performed to select metabolic biomarkers. Finally, a panel of dynamic differential metabolites was used to build logistic regression prediction models.
Results
Metabolite profiles showed a clearly tendency of separation between different follow-up panels. We identified two clusters of 155 serum metabolites with monotonously changing patterns during NCRT (74 decreased metabolites and 81 increased metabolites). Using RM-ANOVA and ML-PLS-DA, 8 metabolites (L-Norleucine, Betaine, Hypoxanthine, Acetylcholine, 1-Hexadecanoyl-sn-glycero-3-phosphocholine, Glycerophosphocholine, Alpha-ketoisovaleric acid, N-Acetyl-L-alanine) were further identified as dynamic differential biomarkers for predicting NCRT sensitivity. The area under the ROC curve (AUC) of prediction model combined with the baseline measurement was 0.54 (95%CI = 0.43 ~ 0.65). By incorporating the variability indexes of 8 dynamic differential metabolites, the prediction model showed better discrimination performance than baseline measurement, with AUC = 0.67 (95%CI 0.57 ~ 0.77), 0.64 (0.53 ~ 0.75), 0.60 (0.50 ~ 0.71), and 0.56 (0.45 ~ 0.67) for the variability index of difference, linear slope, ratio, and standard deviation, respectively.
Conclusion
This study identified eight metabolites as dynamic differential biomarkers to discriminate NCRT-sensitive and resistant patients. The changes of metabolite level during NCRT show better performance in predicting NCRT sensitivity. These findings highlight the clinical significance of metabolites variabilities in metabolomics analysis.



Similar content being viewed by others
Data availability
Metabolomics dataset was available at https://github.com/jiali1021/LARC-dynamic-metabolomics. Other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- TRG:
-
Tumor regression grade
- RSD:
-
Relative standard deviation
- FCM:
-
Fuzzy C-means clustering
- CoA:
-
Coenzyme A
- Ach :
-
Acetylcholine
- 5-FU :
-
5-Fluorouracil
- VIP:
-
Variable importance in projection
- AUC:
-
Area under the receiver operating characteristic curve
- LARC:
-
Locally advanced rectal cancer
- NCRT:
-
Preoperative neoadjuvant chemoradiation therapy
- UHPLC-QTOF-MS:
-
Ultra-high performance liquid chromatography to quadrupole time-of-flight mass spectrometry
- MSCA:
-
Multilevel simultaneous component analysis
- RM-ANOVA:
-
Repeated measure analysis of variance
- ML-PLS-DA:
-
Multilevel partial least-squares discriminant analysis
- KEGG :
-
Kyoto encyclopedia of genes and genomes
- SMPDB:
-
The small molecule pathway database
References
Adam, M. G., et al. (2021). Identification and validation of a multivariable prediction model based on blood plasma and serum metabolomics for the distinction of chronic pancreatitis subjects from non-pancreas disease control subjects. Gut, 70(11), 2150–2158. https://doi.org/10.1136/gutjnl-2020-320723
Aglago, E. K., et al. (2021). Dietary intake and plasma phospholipid concentrations of saturated, monounsaturated and trans fatty acids and colorectal cancer risk in the European prospective investigation into cancer and nutrition cohort. International Journal of Cancer. https://doi.org/10.1002/ijc.33615
Andrade, V. A., et al. (2016). Neoadjuvant therapy and surgery for rectal cancer. Comparative Study between Partial and Complete Pathological Response. Arquivos De Gastroenterologia, 53(3), 163–168. https://doi.org/10.1590/S0004-28032016000300008
Arumugam, M. K., et al. (2021). Beneficial effects of betaine: a comprehensive review. Biology, 10(6), 456. https://doi.org/10.3390/biology10060456
Budhathoki, S., et al. (2017). Association of plasma concentrations of branched-chain amino acids with risk of colorectal adenoma in a large Japanese population. Annals of Oncology: Official Journal of the European Society for Medical Oncology, 28(4), 818–823. https://doi.org/10.1093/annonc/mdw680
Burrage, L. C., et al. (2014). Branched-chain amino acid metabolism: from rare mendelian diseases to more common disorders. Human Molecular Genetics, 23(R1), R1–R8. https://doi.org/10.1093/hmg/ddu123
Chen, G., et al. (2021). Postoperative circulating tumor DNA as markers of recurrence risk in stages II to III colorectal cancer. Journal of Hematology & Oncology, 14(1), 80. https://doi.org/10.1186/s13045-021-01089-z
Cheng, K., et al. (2008). Acetylcholine release by human colon cancer cells mediates autocrine stimulation of cell proliferation. American Journal of Physiology. Gastrointestinal and Liver Physiology, 295(3), G591–G597. https://doi.org/10.1152/ajpgi.00055.2008
Dayde, D., et al. (2017). Predictive and prognostic molecular biomarkers for response to neoadjuvant chemoradiation in rectal cancer. International Journal of Molecular Sciences, 18(3), 573. https://doi.org/10.3390/ijms18030573
Debik, J., et al. (2019). Assessing treatment response and prognosis by serum and tissue metabolomics in breast cancer patients. Journal of Proteome Research, 18(10), 3649–3660. https://doi.org/10.1021/acs.jproteome.9b00316
DeVito, S., et al. (2017). Mutagenic potential of hypoxanthine in live human cells. Mutation Research, 803–805, 9–16. https://doi.org/10.1016/j.mrfmmm.2017.06.005
Fujigaki, S., et al. (2018). Identification of serum biomarkers of chemoradiosensitivity in esophageal cancer via the targeted metabolomics approach. Biomarkers in Medicine, 12(8), 827–840. https://doi.org/10.2217/bmm-2017-0449
Gunderson, L. L., et al. (2010). Revised tumor and node categorization for rectal cancer based on surveillance, epidemiology, and end results and rectal pooled analysis outcomes. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 28(2), 256–263. https://doi.org/10.1200/JCO.2009.23.9194
Hajiasgharzadeh, K., et al. (2020). The dual role of alpha7 nicotinic acetylcholine receptor in inflammation-associated gastrointestinal cancers. Heliyon, 6(3), e03611. https://doi.org/10.1016/j.heliyon.2020.e03611
Jansen, J. J., et al. (2004). Analysis of longitudinal metabolomics data. Bioinformatics (oxford, England), 20(15), 2438–2446. https://doi.org/10.1093/bioinformatics/bth268
Jia, H., et al. (2018). Predicting the pathological response to neoadjuvant chemoradiation using untargeted metabolomics in locally advanced rectal cancer. Radiotherapy and Oncology: Journal of the European Society for Therapeutic Radiology and Oncology, 128(3), 548–556. https://doi.org/10.1016/j.radonc.2018.06.022
Kim, K., et al. (2015). Identification of hypoxanthine and phosphoenolpyruvic acid as serum markers of chemoradiotherapy response in locally advanced rectal cancer. Cancer Research and Treatment, 47(1), 78–89. https://doi.org/10.4143/crt.2013.127
Kuremsky, J. G., et al. (2009). Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. International Journal of Radiation Oncology, Biology, Physics, 74(3), 673–688. https://doi.org/10.1016/j.ijrobp.2009.03.003
Liang, L., et al. (2020). Metabolic dynamics and prediction of gestational age and time to delivery in pregnant women. Cell, 181(7), 1680-1692.e15. https://doi.org/10.1016/j.cell.2020.05.002
Liquet, B., et al. (2012). A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinformatics, 13, 325. https://doi.org/10.1186/1471-2105-13-325
Long, Y., et al. (2017). Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer, 123(20), 4066–4074. https://doi.org/10.1002/cncr.30829
Mason, S. E., et al. (2021). Lipidomic profiling of colorectal lesions for real-time tissue recognition and risk-stratification using rapid evaporative ionisation mass spectrometry. Annals of Surgery. https://doi.org/10.1097/SLA.0000000000005164
Ng, J. W., et al. (2012). The role of longitudinal cohort studies in epigenetic epidemiology: Challenges and opportunities. Genome Biology, 13(6), 246. https://doi.org/10.1186/gb-2012-13-6-246
Onaitis, M. W., et al. (2001). Neoadjuvant chemoradiation for rectal cancer: analysis of clinical outcomes from a 13-year institutional experience. Annals of Surgery, 233(6), 778–785. https://doi.org/10.1097/00000658-200106000-00007
Pakiet, A., et al. (2021). Alterations in complex lipids in tumor tissue of patients with colorectal cancer. Lipids in Health and Disease, 20(1), 85. https://doi.org/10.1186/s12944-021-01512-x
Pang, Z., et al. (2021). MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Research, 49(W1), W388–W396. https://doi.org/10.1093/nar/gkab382
Ryan, J. E., et al. (2015). Assessing pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a systematic review. Colorectal Disease: THe Official Journal of the Association of Coloproctology of Great Britain and Ireland, 17(10), 849–861. https://doi.org/10.1111/codi.13081
Siegel, R. L., et al. (2021). Cancer Statistics 2021. CA: a Cancer Journal for Clinicians, 71(1), 7–33. https://doi.org/10.3322/caac.21654
Smilde, A. K., et al. (2010). Dynamic metabolomic data analysis: a tutorial review. Metabolomics: Official Journal of the Metabolomic Society, 6(1), 3–17. https://doi.org/10.1007/s11306-009-0191-1
Smith, A. M., et al. (2020). A metabolomics approach to screening for autism risk in the children’s autism metabolome project. Autism Research: Official Journal of the International Society for Autism Research, 13(8), 1270–1285. https://doi.org/10.1002/aur.2330
Tian, Y., et al. (2013). Chemopreventive effect of dietary glutamine on colitis-associated colon tumorigenesis in mice. Carcinogenesis, 34(7), 1593–1600. https://doi.org/10.1093/carcin/bgt088
Ueland, P. M. (2011). Choline and betaine in health and disease. Journal of Inherited Metabolic Disease, 34(1), 3–15. https://doi.org/10.1007/s10545-010-9088-4
Valentini, V., et al. (2009). Multidisciplinary rectal cancer management: 2nd european rectal cancer consensus conference (EURECA-CC2). Radiotherapy and Oncology: Journal of the European Society for Therapeutic Radiology and Oncology, 92(2), 148–163. https://doi.org/10.1016/j.radonc.2009.06.027
van de Velde, C. J. H., et al. (2013). EURECCA colorectal: multidisciplinary mission statement on better care for patients with colon and rectal cancer in Europe. European Journal of Cancer (oxford, England: 1990), 49(13), 2784–2790. https://doi.org/10.1016/j.ejca.2013.04.032
van de Velde, C. J. H., et al. (2014). EURECCA colorectal: multidisciplinary management: European consensus conference colon & rectum. European Journal of Cancer (oxford, England: 1990), 50(1), 1.e1-1.e34. https://doi.org/10.1016/j.ejca.2013.06.048
Westerhuis, J. A., et al. (2010). Multivariate paired data analysis: multilevel PLSDA versus OPLSDA. Metabolomics, 6(1), 119–128. https://doi.org/10.1007/s11306-009-0185-z
Xie, G., et al. (2016). Muscarinic receptor signaling and colon cancer progression. Journal of Cancer Metastasis and Treatment, 2, 195–200. https://doi.org/10.20517/2394-4722.2016.05
Youn, J., et al. (2019). Association of choline and betaine levels with cancer incidence and survival: a meta-analysis. Clinical Nutrition (edinburgh, Scotland), 38(1), 100–109. https://doi.org/10.1016/j.clnu.2018.01.042
Yu, J., et al. (2021). Lipidomics and transcriptomics analyses of altered lipid species and pathways in oxaliplatin-treated colorectal cancer cells. Journal of Pharmaceutical and Biomedical Analysis, 200, 114077. https://doi.org/10.1016/j.jpba.2021.114077
Zhang, F., et al. (2017). Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: a systematic review. Oncotarget, 8(21), 35460–35472. https://doi.org/10.18632/oncotarget.16727
Zhang, Y., et al. (2020). Alteration of plasma metabolites associated with chemoradiosensitivity in esophageal squamous cell carcinoma via untargeted metabolomics approach. BMC Cancer, 20(1), 835. https://doi.org/10.1186/s12885-020-07336-9
Zhou, H., et al. (2018). Expression and significance of autonomic nerves and alpha9 nicotinic acetylcholine receptor in colorectal cancer. Molecular Medicine Reports, 17(6), 8423–8431. https://doi.org/10.3892/mmr.2018.8883
Acknowledgements
This study is a joint effort of many investigators and staff members whose contribution is gratefully acknowledged. We appreciate the enrolled adult volunteers for the provision of data.
Funding
This study was supported by grants from Natural Science Foundation of Shanghai (19ZR1410600 [J. Zhu]), Key Research foundation of Zhejiang (2022C03015 [J. Zhu]), National Natural Science Foundation of China (82003554, [H. Jia]), National Natural Science Foundation of China (81973147, 82222064, [T. Zhang]), National Nature Science Foundation of China ([82173613, [Z. Wu]), Scientific Project of Shanghai Municipal Health Commission (201940151, [H. Jia]). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
JL., HJ., JZ. and TZ. designed this study. HJ., JL., MM., JY., ZW., SZ., ZF., BG., BF. and CL. collected clinical samples and clinical information and generated metabolic data. JL. and HJ. conducted data analysis. JL., HJ., JZ. and TZ. wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
No conflicts of interest relevant to this article were reported.
Ethical approval and consent to participate.
Study protocols were approved by the institutional review board of Fudan University Shanghai Cancer Center and written informed consent was obtained from all participants involved in this study.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lv, J., Jia, H., Mo, M. et al. Changes of serum metabolites levels during neoadjuvant chemoradiation and prediction of the pathological response in locally advanced rectal cancer. Metabolomics 18, 99 (2022). https://doi.org/10.1007/s11306-022-01959-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-022-01959-8