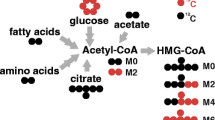
Abstract
Introduction
Carbon isotope tracers have been used to determine relative rates of tricarboxylic acid cycle (TCA) cycle pathways since the 1950s. Steady-state experimental data are typically fit to a single mathematical model of metabolism to determine metabolic fluxes. Whether the chosen model is appropriate for the biological system has generally not been evaluated systematically. An overly-simple model omits known pathways while an overly-complex model may produce incorrect results due to overfitting.
Objectives
The objectives were to develop and study a method that systematically evaluates multiple TCA cycle mathematical models as part of the fitting process.
Methods
The problem of choosing overly-simple or overly-complex models was approached by developing software that automatically explores all possible combinations of flux through pyruvate dehydrogenase, pyruvate kinase, pyruvate carboxylase and anaplerosis at propionyl-CoA carboxylase, and equivalent pathways, all relative to TCA cycle flux. Typical TCA cycle metabolic tracer experiments that use 13C nuclear magnetic resonance for detection and quantification of 13C-enriched glutamate products were simulated and analyzed. By evaluating the multiple model fits with both the conventional sum-of-squares residual error (SSRE) and the Akaike Information Criterion (AIC), the software helps the investigator understand the interaction between model complexity and goodness of fit.
Results
When fitting alternative models of the TCA cycle metabolism, the SSRE may identify more than one model that fits the data well. Among those models, the AIC provides guidance as to which is the simplest of the candidate models is sufficient to describe the observed data. However under some conditions, AIC used alone inappropriately discriminates against necessary metabolic complexity.
Conclusion
In combination, the SSRE and AIC help the investigator identify the model that best describes the metabolism of a biological system.






Similar content being viewed by others
Code availability
The MATLAB source code of software described in this article is curated on GitHub. Contact the corresponding author for download permission.
References
Akaike, H. (1974). A new look at the statistical model identification. IEEE Transactions on Automatic Control 19, 716–723 https://doi.org/10.1109/tac.1974.1100705
Alger, J. R., Sherry, A. D., & Malloy, C. R. (2018). tcaSIM: A simulation program for optimal design of (13)C tracer experiments for analysis of metabolic flux by NMR and mass spectroscopy. Current Metabolomics, 6, 176–187. https://doi.org/10.2174/2213235X07666181219115856.
Anderson, D. R. (2008). Model based inference in the life sciences: a primer on evidence. New York, London: Springer.
Antoniewicz, M. R., Kelleher, J. K., & Stephanopoulos, G. (2006). Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metabolic Engineering, 8, 324–37. https://doi.org/10.1016/j.ymben.2006.01.004.
Banwarth-Kuhn, M., & Sindi, S. (2020). How and why to build a mathematical model: A case study using prion aggregation. Journal of Biological Chemistry, 295, 5022–5035. https://doi.org/10.1074/jbc.REV119.009851.
Chen, Y., et al. (2011). Longitudinal regression analysis of spatial-temporal growth patterns of geometrical diffusion measures in early postnatal brain development with diffusion tensor imaging. Neuroimage, 58, 993–1005. https://doi.org/10.1016/j.neuroimage.2011.07.006.
Cohen, S. M. (1987). Effects of insulin on perfused liver from streptozotocin-diabetic and untreated rats: 13 C NMR assay of pyruvate kinase flux. Biochemistry, 26, 573–80. https://doi.org/10.1021/bi00376a032.
D’Errico, J. (2006). fminsearchbnd (version 1.4.0.0). MathWorks File Exchange. Retrieved from https://www.mathworks.com/matlabcentral/fileexchange/8277-fminsearchbnd-fminsearchcon
Deja, S., et al. (2020). Simultaneous tracers and a unified model of positional and mass isotopomers for quantification of metabolic flux in liver. Metabolic Engineering, 59, 1–14. https://doi.org/10.1016/j.ymben.2019.12.005.
Ferizi, U., et al. (2015). White matter compartment models for in vivo diffusion MRI at 300mT/m. Neuroimage, 118, 468–483. https://doi.org/10.1016/j.neuroimage.2015.06.027.
Fernandez, C. A., & Des Rosiers, C. (1995). Modeling of liver citric acid cycle and gluconeogenesis based on 13 C mass isotopomer distribution analysis of intermediates. Journal of Biological Chemistry, 270, 10037–10042. https://doi.org/10.1074/jbc.270.17.10037.
Freidmann, B., Goodman, E. H., Jr., Saunders, H. L., Kostos, V., & Weinhouse, S. (1971). An estimation of pyruvate recycling during gluconeogenesis in the perfused rat liver. Archives of Biochemistry and Biophysics, 143, 566–578. https://doi.org/10.1016/0003-9861(71)90241-4.
Friedman, B., Goodman, E. H., Jr., Saunders, H. L., Kostos, V., & Weinhouse, S. (1971). Estimation of pyruvate recycling during gluconeogenesis in perfused rat liver. Metabolism: Clinical and Experimental, 20, 2–12. https://doi.org/10.1016/0026-0495(71)90055-2.
Jeffrey, F. M., Storey, C. J., Sherry, A. D., & Malloy, C. R. (1996). 13 C isotopomer model for estimation of anaplerotic substrate oxidation via acetyl-CoA. American Journal of Physiology, 271, E788–E799. https://doi.org/10.1152/ajpendo.1996.271.4.E788.
Jin, H., & Moseley, H. N. B. (2020). Robust moiety model selection using mass spectrometry measured isotopologues. Metabolites. https://doi.org/10.3390/metabo10030118.
Jones, J. G., Naidoo, R., Sherry, A. D., Jeffrey, F. M., Cottam, G. L., & Malloy, C. R. (1997). Measurement of gluconeogenesis and pyruvate recycling in the rat liver: A simple analysis of glucose and glutamate isotopomers during metabolism of [1,2,3-(13)C3]propionate. FEBS Letters, 412, 131–137. https://doi.org/10.1016/s0014-5793(97)00764-3.
Katz, J. (1985). Determination of gluconeogenesis in vivo with 14 C-labeled substrates. American Journal of Physiology, 248, R391–R399. https://doi.org/10.1152/ajpregu.1985.248.4.R391.
Konishi, S., & Kitagawa, G. (1996). Generalised information criteria in model selection. Biometrika, 83, 875–890.
Kornberg, H. L. (1966). Anaplerotic sequences and their role in metabolism. Essays in Biochemistry, 2, 1–31.
Lipkin, E. W., Teller, D. C., & de Haen, C. (1986). Equilibrium binding of insulin to rat white fat cells at 15 degrees C. Journal of Biological Chemistry, 261, 1694–1701.
Lipkin, E. W., Teller, D. C., & de Haen, C. (1986). Kinetics of insulin binding to rat white fat cells at 15 degrees C. Journal of Biological Chemistry, 261, 1702–1711.
Magnusson, I., et al. (1991). Noninvasive tracing of Krebs cycle metabolism in liver. Journal of Biological Chemistry, 266, 6975–6984.
Malloy, C. R., Maher, E. H., Marin-Valencia, I., Mickey, B., DeBerardinis, R. J., & Sherry, A. D. (2013). Carbon-13 nuclear magnetic resonance for analysis of metabolic pathways. In N. W. Lutz, J. V. Sweedler, & R. A. Wevers (Eds.), Methodologies for metabolomics: Experimental strategies and techniques (pp. 415–445). London: Cambridge University Press.
Malloy, C. R., Sherry, A. D., & Jeffrey, F. M. (1988). Evaluation of carbon flux and substrate selection through alternate pathways involving the citric acid cycle of the heart by 13 C NMR spectroscopy. Journal of Biological Chemistry, 263, 6964–6971.
Malloy, C. R., Sherry, A. D., & Jeffrey, F. M. (1990). Analysis of tricarboxylic acid cycle of the heart using 13 C isotope isomers. American Journal of Physiology, 259, H987–H995. https://doi.org/10.1152/ajpheart.1990.259.3.H987.
Malloy, C. R., Thompson, J. R., Jeffrey, F. M., & Sherry, A. D. (1990). Contribution of exogenous substrates to acetyl coenzyme A: Measurement by 13 C NMR under non-steady-state conditions. Biochemistry, 29, 6756–6761. https://doi.org/10.1021/bi00481a002.
Mashimo, T., et al. (2014). Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell, 159, 1603–1614. https://doi.org/10.1016/j.cell.2014.11.025.
Mason, G. F., Rothman, D. L., Behar, K. L., & Shulman, R. G. (1992). NMR determination of the TCA cycle rate and alpha-ketoglutarate/glutamate exchange rate in rat brain. Journal of Cerebral Blood Flow and Metabolism, 12, 434–447. https://doi.org/10.1038/jcbfm.1992.61.
Nuutinen, E. M., Peuhkurinen, K. J., Pietilainen, E. P., Hiltunen, J. K., & Hassinen, I. E. (1981). Elimination and replenishment of tricarboxylic acid-cycle intermediates in myocardium. Biochemical Journal, 194, 867–875. https://doi.org/10.1042/bj1940867.
Owen, O. E., Kalhan, S. C., & Hanson, R. W. (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. Journal of Biological Chemistry, 277, 30409–30412. https://doi.org/10.1074/jbc.R200006200.
Perry, R. J., et al. (2017). Non-invasive assessment of hepatic mitochondrial metabolism by positional isotopomer NMR tracer analysis (PINTA). Nat Commun, 8, 798. https://doi.org/10.1038/s41467-017-01143-w.
Petersen, K. F., Blair, J. B., & Shulman, G. I. (1995). Triiodothyronine treatment increases substrate cycling between pyruvate carboxylase and malic enzyme in perfused rat liver. Metabolism: Clinical and Experimental, 44, 1380–1383. https://doi.org/10.1016/0026-0495(95)90133-7.
Petersen, K. F., Dufour, S., Cline, G. W., & Shulman, G. I. (2019). Regulation of hepatic mitochondrial oxidation by glucose-alanine cycling during starvation in humans. Journal of Clinical Investigation, 129, 4671–4675. https://doi.org/10.1172/JCI129913.
Peuhkurinen, K. J., & Hassinen, I. E. (1982). Pyruvate carboxylation as an anaplerotic mechanism in the isolated perfused rat heart. Biochemical Journal, 202, 67–76. https://doi.org/10.1042/bj2020067.
Peuhkurinen, K. J., Nuutinen, E. M., Pietilainen, E. P., Hiltunen, J. K., & Hassinen, I. E. (1982). Role of pyruvate carboxylation in the energy-linked regulation of pool sizes of tricarboxylic acid-cycle intermediates in the myocardium. Biochemical Journal, 208, 577–581. https://doi.org/10.1042/bj2080577.
Rabinowitz, J. D., & Vastag, L. (2012). Teaching the design principles of metabolism. Nature Chemical Biology, 8, 497–501. https://doi.org/10.1038/nchembio.969.
Rognstad, R. (1979). Pyruvate cycling involving possible oxaloacetate decarboxylase activity. Biochimica et Biophysica Acta, 586, 242–249. https://doi.org/10.1016/0304-4165(79)90096-5.
Rognstad, R., & Katz, J. (1972). Gluconeogenesis in the kidney cortex. Quantitative estimation of carbon flow. Journal of Biological Chemistry, 247, 6047–6054.
Samoilov, M., Plyasunov, S., & Arkin, A. P. (2005). Stochastic amplification and signaling in enzymatic futile cycles through noise-induced bistability with oscillations. Proceedings of the National Academy of Sciences of the United States of America, 102, 2310–2315. https://doi.org/10.1073/pnas.0406841102.
Schumann, W. C., Magnusson, I., Chandramouli, V., Kumaran, K., Wahren, J., & Landau, B. R. (1991). Metabolism of [2-14 C] acetate and its use in assessing hepatic Krebs cycle activity and gluconeogenesis. Journal of Biological Chemistry, 266, 6985–6990.
Sherry, A. D., Jeffrey, F. M., & Malloy, C. R. (2004). Analytical solutions for (13)C isotopomer analysis of complex metabolic conditions: substrate oxidation, multiple pyruvate cycles, and gluconeogenesis. Metabolic Engineering, 6, 12–24. https://doi.org/10.1016/j.ymben.2003.10.007.
Sherry, A. D., Malloy, C. R., Roby, R. E., Rajagopal, A., & Jeffrey, F. M. (1988). Propionate metabolism in the rat heart by 13 C n.m.r. spectroscopy. Biochemical Journal, 254, 593–598. https://doi.org/10.1042/bj2540593.
Sherry, A. D., Malloy, C. R., Zhao, P., & Thompson, J. R. (1992). Alterations in substrate utilization in the reperfused myocardium: a direct analysis by 13 C NMR. Biochemistry, 31, 4833–4837. https://doi.org/10.1021/bi00135a014.
Shestov, A. A., et al. (2016). Bonded cumomer analysis of human melanoma metabolism monitored by 13 C NMR spectroscopy of perfused tumor cells. Journal of Biological Chemistry, 291, 5157–5171. https://doi.org/10.1074/jbc.M115.701862.
Strisower, E. H., Kohler, G. D., & Chaikoff, I. L. (1952). Incorporation of acetate carbon into glucose by liver slices from normal and alloxan-diabetic rats. Journal of Biological Chemistry, 198, 115–126.
Sundqvist, K. E., Peuhkurinen, K. J., Hiltunen, J. K., & Hassinen, I. E. (1984). Effect of acetate and octanoate on tricarboxylic acid cycle metabolite disposal during propionate oxidation in the perfused rat heart. Biochimica et Biophysica Acta, 801, 429–436. https://doi.org/10.1016/0304-4165(84)90149-1.
Taegtmeyer, H., Hems, R., & Krebs, H. A. (1980). Utilization of energy-providing substrates in the isolated working rat heart. Biochemical Journal, 186, 701–711. https://doi.org/10.1042/bj1860701.
Weinman, E. O., Strisower, E. H., & Chaikoff, I. L. (1957). Conversion of fatty acids to carbohydrate; application of isotopes to this problem and role of the Krebs cycle as a synthetic pathway. Physiological Reviews, 37, 252–272. https://doi.org/10.1152/physrev.1957.37.2.252.
Wright, B. E., & Kelly, P. J. (1981). Kinetic models of metabolism in intact cells, tissues, and organisms. Current Topics in Cellular Regulation, 19, 103–158. https://doi.org/10.1016/b978-0-12-152819-5.50021-x.
Yang, C., et al. (2014). Simultaneous steady-state and dynamic 13 C NMR can differentiate alternative routes of pyruvate metabolism in living cancer cells. Journal of Biological Chemistry, 289, 6212–6224. https://doi.org/10.1074/jbc.M113.543637.
Yang, J., Kalhan, S. C., & Hanson, R. W. (2009). What is the metabolic role of phosphoenolpyruvate carboxykinase? Journal of Biological Chemistry, 284, 27025–27029. https://doi.org/10.1074/jbc.R109.040543.
Young, J. D. (2014). INCA: A computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics, 30, 1333–1335. https://doi.org/10.1093/bioinformatics/btu015.
Funding
This study was supported by National Institutes of Health Grants DK058398, HL034557 and EB015908. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
JRA and CRM conceived and designed the research. JRA conducted experiments. JRA and CRM analyzed data. AM provided biostatistical expertise. ADS provided background knowledge. JRA and CRM wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest in regards to this manuscript.
Ethical approval
This article does not contain any studies with human and/or animal participants performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alger, J.R., Minhajuddin, A., Dean Sherry, A. et al. Analysis of steady-state carbon tracer experiments using akaike information criteria. Metabolomics 17, 61 (2021). https://doi.org/10.1007/s11306-021-01807-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-021-01807-1