Abstract
Introduction
Metabolomics is a promising approach for discovery of relevant biomarkers in cells, tissues, organs, and biofluids for disease identification and prediction. The field has mostly relied on blood-based biofluids (serum, plasma, urine) as non-invasive sources of samples as surrogates of tissue or organ-specific conditions. However, the tissue specificity of metabolites pose challenges in translating blood metabolic profiles to organ-specific pathophysiological changes, and require further downstream analysis of the metabolites.
Objectives
As part of this project, we aim to develop and optimize an efficient extraction protocol for the analysis of kidney tissue metabolites representative of key primate metabolic pathways.
Methods
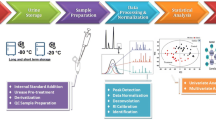
Kidney cortex and medulla tissues of a baboon were homogenized and extracted using eight different extraction protocols including methanol/water, dichloromethane/methanol, pure methanol, pure water, water/methanol/chloroform, methanol/chloroform, methanol/acetonitrile/water, and acetonitrile/isopropanol/water. The extracts were analyzed by a two-dimensional gas chromatography time-of-flight mass-spectrometer (2D GC–ToF-MS) platform after methoximation and silylation.
Results
Our analysis quantified 110 shared metabolites in kidney cortex and medulla tissues from hundreds of metabolites found among the eight different solvent extractions spanning low to high polarities. The results revealed that medulla is metabolically richer compared to the cortex. Dichloromethane and methanol mixture (3:1) yielded highest number of metabolites across both the tissue types. Depending on the metabolites of interest, tissue type, and the biological question, different solvents can be used to extract specific groups of metabolites.
Conclusion
This investigation provides insights into selection of extraction solvents for detection of classes of metabolites in renal cortex and medulla, which is fundamentally important for identification of prognostic and diagnostic metabolic kidney biomarkers for future therapeutic applications.





Similar content being viewed by others
References
Arthur, J. M., Thongboonkerd, V., Scherzer, J. A., Cai, J., Pierce, W. M., & Klein, J. B. (2002). Differential expression of proteins in renal cortex and medulla: A proteomic approach. Kidney International, 62(4), 1314–1321. https://doi.org/10.1111/j.1523-1755.2002.kid588.x.
Boudonck, K. J., Mitchell, M. W., Német, L., Keresztes, L., Nyska, A., Shinar, D., & Rosenstock, M. (2009). Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicologic Pathology, 37(3), 280–292. https://doi.org/10.1177/0192623309332992.
Caraux, G., & Pinloche, S. (2005). PermutMatrix: A graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics, 21, 1280–1281. https://doi.org/10.1093/bioinformatics/bti141.
Chambers, J. M. (2017). Graphical methods for data analysis. Boca Raton: CRC Press.
Chen, S., Hoene, M., Li, J., Li, Y., Zhao, X., Häring, H. U., Schleicher, E. D., Weigert, C., Xu, G., & Lehmann, R. (2013). Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. Journal of Chromatography A, 1298, 9–16. https://doi.org/10.1016/j.chroma.2013.05.019.
Connor, S. C., Hansen, M. K., Corner, A., Smith, R. F., & Ryan, T. E. (2010). Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Molecular BioSystems, 6(5), 909–921. https://doi.org/10.1039/B914182K.
Conway, J. R., Lex, A., & Gehlenborg, N. (2017). UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics, 33(18), 2938–2940. https://doi.org/10.1093/bioinformatics/btx364.
Cox, L. A., Comuzzie, A. G., Havill, L. M., Karere, G. M., Spradling, K. D., Mahaney, M. C. et al. (2013). Baboons as a model to study genetics and epigenetics of human disease. ILAR Journal, 54(2), 106–121.
Fiehn, O. (2016). Metabolomics by gas chromatography–mass spectrometry: Combined targeted and untargeted profiling. Current Protocols in Molecular Biology. https://doi.org/10.1002/0471142727.mb3004s114.
Fiehn, O., Wohlgemuth, G., & Scholz, M. (2005). Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. In B. Ludäscher & L. Raschid (Eds.), Data integration in the life sciences. DILS 2005. Lecture notes in computer science (Vol. 3615). Berlin: Springer. https://doi.org/10.1007/11530084_18.
Gebhard, R. L., Clayman, R. V., Prigge, W. F., Figenshau, R., Staley, N. A., Reesey, C., & Bear, A. (1987). Abnormal cholesterol metabolism in renal clear cell carcinoma. Journal of Lipid Research, 28(10), 1177–1184.
Grapov, D. (2014) DeviumWeb: Version 0.3.2. ZENODO. https://doi.org/10.5281/zenodo.12879, https://github.com/dgrapov/DeviumWeb.
Hallan, S., Afkarian, M., Zelnick, L. R., Kestenbaum, B., Sharma, S., Saito, R., Darshi, M., Barding, G., Raftery, D., Ju, W., & Kretzler, M. (2017). Metabolomics and gene expression analysis reveal down-regulation of the citric acid (TCA) cycle in non-diabetic CKD patients. EBioMedicine, 26, 68–77. https://doi.org/10.1016/j.ebiom.2017.10.027.
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., Ojima, Y., Tanaka, K., Tanaka, S., Aoshima, K., & Oda, Y. (2010). MassBank: A public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry, 45(7), 703–714. https://doi.org/10.1002/jms.1777.
Ibáñez, C., Simó, C., Palazoglu, M., & Cifuentes, A. (2017). GC-MS based metabolomics of colon cancer cells using different extraction solvents. Analytica Chimica Acta, 986, 48–56. https://doi.org/10.1016/j.aca.2017.07.019.
Karlsson, K. A., Samuelsson, B. E., & Steen, G. O. (1973). The sphingolipid composition of bovine kidney cortex, medulla and papilla. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism, 316(3), 317–335.
Kim, K., Aronov, P., Zakharkin, S. O., Anderson, D., Perroud, B., Thompson, I. M., & Weiss, R. H. (2009). Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Molecular & Cellular Proteomics, 8(3), 558–570. https://doi.org/10.1074/mcp.M800165-MCP200.
Kind, T., Tolstikov, V., Fiehn, O., & Weiss, R. H. (2007). A comprehensive urinary metabolomic approach for identifying kidney cancer. Analytical Biochemistry, 363(2), 185–195. https://doi.org/10.1016/j.ab.2007.01.028.
Kopka, J., Schauer, N., Krueger, S., Birkemeyer, C., Usadel, B., Bergmüller, E., et al. (2005). GMD@ CSB. DB: The Golm metabolome database. Bioinformatics, 21, 1635–1638. https://doi.org/10.1093/bioinformatics/bti236.
Lee, J. W., Chou, C. L., & Knepper, M. A. (2015). Deep sequencing in microdissected renal tubules identifies nephron segment–specific transcriptomes. Journal of the American Society of Nephrology. https://doi.org/10.1681/ASN.2014111067.
Leuthold, P., Schaeffeler, E., Winter, S., Büttner, F., Hofmann, U., Mürdter, T. E., Rausch, S., Sonntag, D., Wahrheit, J., Fend, F., & Hennenlotter, J. (2017). Comprehensive metabolomic and lipidomic profiling of human kidney tissue: A platform comparison. Journal of Proteome Research, 16(2), 933–944. https://doi.org/10.1021/acs.jproteome.6b00875.
Lin, C. Y., Wu, H., Tjeerdema, R. S., & Viant, M. R. (2007). Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics, 3(1), 55–67. https://doi.org/10.1007/s11306-006-0043-1.
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., & Fernie, A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols, 1, 387–396. https://doi.org/10.1038/nprot.2006.59.
Little, J. L. (1999). Artifacts in trimethylsilyl derivatization reactions and ways to avoid them. Journal of Chromatography A, 844(1), 1–22. https://doi.org/10.1016/S0021-9673(99)00267-8.
Mahadevan, S., Shah, S. L., Marrie, T. J., & Slupsky, C. M. (2008). Analysis of metabolomic data using support vector machines. Analytical Chemistry, 80(19), 7562–7570. https://doi.org/10.1021/ac800954c.
Martin, A. C., Pawlus, A. D., Jewett, E. M., Wyse, D. L., Angerhofer, C. K., & Hegeman, A. D. (2014). Evaluating solvent extraction systems using metabolomics approaches. RSC Advances, 4(50), 26325–26334. https://doi.org/10.1039/C4RA02731K.
Masson, P., Alves, A. C., Ebbels, T. M., Nicholson, J. K., & Want, E. J. (2010). Optimization and evaluation of metabolite extraction protocols for untargeted metabolic profiling of liver samples by UPLC-MS. Analytical Chemistry, 82(18), 7779–7786. https://doi.org/10.1021/ac101722e.
Masson, P., Spagou, K., Nicholson, J. K., & Want, E. J. (2011). Technical and biological variation in UPLC–MS-based untargeted metabolic profiling of liver extracts: Application in an experimental toxicity study on galactosamine. Analytical Chemistry, 83(3), 1116–1123. https://doi.org/10.1021/ac103011b.
Melnick, J. Z., Baum, M., & Thompson, J. R. (1994). Aminoglycoside-induced Fanconi’s syndrome. American Journal of Kidney Diseases, 23(1), 118–122. https://doi.org/10.1016/S0272-6386(12)80820-1.
Nielsen, P. M., Eldirdiri, A., Bertelsen, L. B., Jørgensen, H. S., Ardenkjaer-Larsen, J. H., & Laustsen, C. (2017). Fumarase activity: An in vivo and in vitro biomarker for acute kidney injury. Scientific Reports. https://doi.org/10.1038/srep40812.
Pastore, A., Noce, A., Di Giovamberardino, G., De Stefano, A., Callà, C., Zenobi, R., Dessì, M., & Di Daniele, N. (2015). Homocysteine, cysteine, folate and vitamin B12 status in type 2 diabetic patients with chronic kidney disease. Journal of Nephrology, 28(5), 571–576. https://doi.org/10.1007/s40620-014-0126-4.
Patterson, A. D., Bonzo, J. A., Li, F., Krausz, K. W., Eichler, G. S., Aslam, S., Tigno, X., Weinstein, J. N., Hansen, B. C., Idle, J. R., & Gonzalez, F. J. (2011). Metabolomics reveals attenuation of the SLC6A20 kidney transporter in nonhuman primate and mouse models of type 2 diabetes mellitus. Journal of Biological Chemistry, 286(22), 19511–19522. https://doi.org/10.1074/jbc.M111.221739.
Pickering, C. M., Grady, C., Medvar, B., Emamian, M., Sandoval, P. C., Zhao, Y., Yang, C. R., Jung, H. J., Chou, C. L., & Knepper, M. A. (2016). Proteomic profiling of nuclear fractions from native renal inner medullary collecting duct cells. Physiological Genomics, 48(2), 154–166. https://doi.org/10.1152/physiolgenomics.00090.2015.
R Development Core Team. (2017). R: A language and environment for statistical computing [Internet]. Vienna: R Foundation for Statistical Computing.
Rhee, E. P., Clish, C. B., Wenger, J., Roy, J., Elmariah, S., Pierce, K. A., Bullock, K., Anderson, A. H., Gerszten, R. E., & Feldman, H. I. (2016). Metabolomics of chronic kidney disease progression: A case-control analysis in the chronic renal insufficiency cohort study. American Journal of Nephrology, 43(5), 366–374. https://doi.org/10.1159/000446484.
Sokal, R. R., & Rohlf, F. J. (1995). Biometry: The principles and practice of statistics in biological research. New York: W.H. Freeman and Company.
Sonmez, G., Mutlu, H., Ozturk, E., Sildiroglu, H. O., Keskin, A. T., Basekim, C. C., & Kizilkaya, E. (2007). Magnetic resonance imaging findings of adult-onset glutaric aciduria type I. Acta Radiologica, 48(5), 557–559.
Suhre, K., Meisinger, C., Döring, A., Altmaier, E., Belcredi, P., Gieger, C., Chang, D., Milburn, M. V., Gall, W. E., Weinberger, K. M., & Mewes, H. W. (2010). Metabolic footprint of diabetes: A multiplatform metabolomics study in an epidemiological setting. PLoS ONE, 5(11), e13953. https://doi.org/10.1371/journal.pone.0013953.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., Fan, T. W. M., Fiehn, O., Goodacre, R., Griffin, J. L., & Hankemeier, T. (2007). Proposed minimum reporting standards for chemical analysis. Metabolomics, 3(3), 211–221. https://doi.org/10.1007/s11306-007-0082-2.
Sun, J., Shannon, M., Ando, Y., Schnackenberg, L. K., Khan, N. A., Portilla, D., & Beger, R. D. (2012). Serum metabolomic profiles from patients with acute kidney injury: A pilot study. Journal of Chromatography B, 893, 107–113. https://doi.org/10.1016/j.jchromb.2012.02.042.
Van der Kloet, F. M., Tempels, F. W. A., Ismail, N., Van der Heijden, R., Kasper, P. T., Rojas-Cherto, M., Van Doorn, R., Spijksma, G., Koek, M., Van der Greef, J., & Mäkinen, V. P. (2012). Discovery of early-stage biomarkers for diabetic kidney disease using MS-based metabolomics (FinnDiane study). Metabolomics, 8(1), 109–119. https://doi.org/10.1007/s11306-011-0291-6.
Vorkas, P. A., Isaac, G., Anwar, M. A., Davies, A. H., Want, E. J., Nicholson, J. K., & Holmes, E. (2015). Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: Application to cardiovascular disease. Analytical Chemistry, 87(8), 4184. https://doi.org/10.1021/ac503775m.
Wachsmuth, C. J., Vogl, F. C., Oefner, P. J., & Dettmer, K. (2013). Gas chromatographic techniques in metabolomics. In T. Hyotylainen, S. Wiedmer (Eds.), Chromatographic methods in metabolomics (pp. 87–105).
Weiss, R. H., & Kim, K. (2012). Metabolomics in the study of kidney diseases. Nature Reviews Nephrology, 8(1), 22–33. https://doi.org/10.1038/nrneph.2011.152.
Winnike, J. H., Wei, X., Knagge, K. J., Colman, S. D., Gregory, S. G., & Zhang, X. (2015). Comparison of GC-MS and GC × GC-MS in the analysis of human serum samples for biomarker discovery. Journal of Proteome Research, 14(4), 1810–1817. https://doi.org/10.1021/pr5011923.
Wishart, D. S. (2012). Small molecules and disease. PLoS Computational Biology, 8(12), e1002805. https://doi.org/10.1371/journal.pcbi.1002805.
Wu, H., Southam, A. D., Hines, A., & Viant, M. R. (2008). High-throughput tissue extraction protocol for NMR-and MS-based metabolomics. Analytical Biochemistry, 372(2), 204–212. https://doi.org/10.1016/j.ab.2007.10.002.
Xia, J., Sinelnikov, I. V., Han, B., & Wishart, D. S. (2015). MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Research, 43(W1), W251–W257. https://doi.org/10.1093/nar/gkv380.
You, Y. H., Quach, T., Saito, R., Pham, J., & Sharma, K. (2015). Metabolomics reveals a key role for fumarate in mediating the effects of NADPH oxidase 4 in diabetic kidney disease. Journal of the American Society of Nephrology. https://doi.org/10.1681/ASN.2015030302.
Zukunft, S., Prehn, C., Röhring, C., Möller, G., de Angelis, M. H., Adamski, J., & Tokarz, J. (2018). High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics, 14(1), 18. https://doi.org/10.1007/s11306-017-1312-x.
Acknowledgements
This work was supported by a Forum Grant (BM-17-04629) awarded to BBM by the Texas Biomedical Research Institute, San Antonio, Texas.
Author information
Authors and Affiliations
Contributions
BBM envisioned the project, BBM, and LAC designed the research; BBM and RPU performed the experiments; MO provided essential reagents and materials, BBM analyzed the data, BBM, MO, LAC wrote the manuscript, and BBM interpreted the data, has the primary responsibility for the final content, and edits. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest and no conflicts of interest.
Ethical approval
The baboon kidney samples were collected under IACUC approved protocols at facilities located at the Southwest National Primate Research Center, Texas Biomedical Research Institute, San Antonio, United States. All applicable international, national, and institutional guidelines for the care and use of animals were followed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Misra, B.B., Upadhayay, R.P., Cox, L.A. et al. Optimized GC–MS metabolomics for the analysis of kidney tissue metabolites. Metabolomics 14, 75 (2018). https://doi.org/10.1007/s11306-018-1373-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-018-1373-5