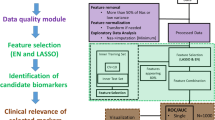
Abstract
Introduction
In the context of metabolomics analyses, partial least squares (PLS) represents the standard tool to perform regression and classification. OPLS, the Orthogonal extension of PLS which has proved to be very useful when interpretation is the main issue, is a more recent way to decompose the PLS solution into predictive components correlated to the target Y and components pertaining to the data X but uncorrelated to Y. This predominance of (O)PLS can raise the question of the awareness of alternative multivariate regression and/or classification tools able to find biomarkers. Actually, the search for biomarkers remains a key issue in metabolomics as it is crucial to very accurately target discriminating features.
Objective
Most of the time, (O)PLS methods perform well but a drawback often occurs: too many variables can be selected as potential biomarkers even using adapted statistical significance tests. However, for final users (in medical studies for instance), it can be advantageous to deal with only a small number of easily interpretable biomarkers.
Methods
This drawback is approached in this paper via the use of sparse methods. The sparse-PLS (sPLS), an extension of PLS which promotes an inner variable/feature selection, is an interesting existing solution. But a new intuitive algorithm is proposed in this paper to combine sparsity and the advantages of an orthogonalization step: the “Light-sparse-OPLS” (L-sOPLS). L-sOPLS promotes sparsity on a previously optimized deflated matrix which implies the removal of the Y-orthogonal components.
Results
A discussion around the compromise between sparsity and predictive modelling performances is provided and it is shown that L-sOPLS produces convincing results, illustrated principally on the basis of \(^1\)H-NMR spectral data but also on genomic RT-qPCR data.
Conclusion
The L-sOPLS algorithm allows to reach better predictive performances than (O)PLS and sPLS while taking into account only a very small number of relevant descriptors.



Similar content being viewed by others
References
Abdi, H. (2010). Partial least squares regression and projection on latent structure regression (pls regression). Wiley Interdisciplinary Reviews: Computational Statistics, 2(1), 97–106.
Afanador, N. L., Tran, T. N., & Buydens, L. (2013). Use of the bootstrap and permutation methods for a more robust variable importance in the projection metric for partial least squares regression. Analytica Chimica Acta, 768, 49–56.
Barker, M., & Rayens, W. (2003). Partial least squares for discrimination. Journal of Chemometrics, 17(3), 166–173.
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136(2), 215–233.
Bylesjo, M., Rantalainen, M., Cloarec, O., & Nicholson, J. (2006). OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics, 20(8–10), 341–351.
Chapman, A., & Saad, Y. (1997). Deflated and augmented Krylov subspace techniques. Numerical Linear Algebra with Applications, 4(1), 43–66.
Chun, H., & Keles, S. (2007). Sparse partial least squares regression with an application to genome scale transcription factor analysis. Madison: Department of Statistics, University of Wisconsin.
Chung, D., Chun, H., & Keles, S. (2012). Spls: Sparse partial least squares (SPLS) regression and classification. R package, version, 2, 1–1.
De Jong, S. (1993). SIMPLS: An alternative approach to partial least squares regression. Chemometrics and Intelligent Laboratory Systems, 18(3), 251–263.
Efron, B., Hastie, T., Johnstone, I., & Tibshirani, R. (2004). Least angle regression. Annals of Statistics, 32(2), 407499.
Feraud, B., Govaerts, B., Verleysen, M., & De Tullio, P. (2015). Statistical treatment of 2D NMR COSY spectra in metabolomics: Data preparation, clustering-based evaluation of the metabolomic informative content and comparison with \(^1\)H-NMR. Metabolomics, 11(6), 1756–1768.
Friedman J., Hastie T., & Tibshirani R. (2010). A note on the group lasso and a sparse group lasso, arXiv preprint arXiv:1001.0736.
Gabrielsson, J., Jonsson, H., Airiaub, C., & Schmidt, B. (2006). OPLS methodology for analysis of pre-processing effects on spectroscopic data. Chemometrics and Intelligent Laboratory Systems, 84(1–2), 153–158.
Geladi, P., & Kowalski, B. R. (1986). Partial least squares regression: A tutorial. Analytica Chimica Acta, 185, 1–17.
Giudice, L. C., & Kao, L. C. (2004). Endometriosis. Lancet, 364, 178999.
Hastie, T., Tibshirani, R., & Wainwright, M. (2015). Statistical learning with sparsity: The lasso and generalizations. Boca Raton: CRC Press.
Hoskuldsson, A. (1988). PLS regression methods. Journal of Chemometrics, 2(3), 211–228.
Indahl, U. G., Liland, K. H., & Ns, T. (2009). Canonical partial least squares: A unified PLS approach to classification and regression problems. Journal of Chemometrics, 23(9), 495–504.
Jung, Y., Lee, J., Kwon, J., Lee, K. S., Ryu, D. H., & Hwang, G. S. (2010). Discrimination of the geographical origin of beef by \(^1\)H-NMR-based metabolomics. Journal of Agricultural and Food Chemistry, 58(19), 10458–10466.
Lai, E. C. (2002). Micro RNAs are complementary to 3 UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genetics, 30, 363.
Lê Cao, K. A., Rossouw, D., Robert-Grani, C., & Besse, P. (2008). A sparse PLS for variable selection when integrating omics data. Statistical Applications in Genetics and Molecular Biology, 7(1), 35.
Lu, B., Castillo, I., Chiang, L., & Edgar, T. F. (2014). Industrial PLS model variable selection using moving window variable importance in projection. Chemometrics and Intelligent Laboratory Systems, 135, 90–109.
Mevik, B. H., & Cederkvist, H. R. (2004). Mean squared error of prediction (MSEP) estimates for principal component regression (PCR) and partial least squares regression (PLSR). Journal of Chemometrics, 18(9), 422–429.
Munoz-Romero, S., Arenas-Garca, J., & Gmez-Verdejo, V. (2015). Sparse and kernel OPLS feature extraction based on eigenvalue problem solving. Pattern Recognition, 48(5), 1797–1811.
Nisenblat V., Bossuyt P. M., Shaikh R., Farquhar C., Jordan V., Scheffers C. S., ... & Hull M. L. (2016). Blood biomarkers for the non-invasive diagnosis of endometriosis. The Cochrane Library.
Rousseau, R. (2011). Statistical contribution to the analysis of metabonomic data in \({}^1\)H-NMR spectroscopy (Doctoral dissertation, Université Catholique de Louvain, Belgium), permalink: http://hdl.handle.net/2078.1/75532.
Stenlund, H., Gorzsas, A., Persson, P., Sundberg, B., & Trygg, J. (2008). Orthogonal projections to latent structures discriminant analysis modeling on in situ FT-IR spectral imaging of liver tissue for identifying sources of variability. Analytical Chemistry, 80(18), 6898–6906.
Tapp, H. S., & Kemsley, E. K. (2009). Notes on the practical utility of OPLS. TrAC Trends in Analytical Chemistry, 28(11), 1322–1327.
Trygg, J., & Wold, S. (2002). Orthogonal projections to latent structures (O-PLS). Journal of Chemometrics, 16(3), 119–128.
van Gerven, M. A. J., & Heskes, T. (2010). Sparse orthonormalized partial least squares. In Benelux conference on artificial intelligence.
Wehrens, R. (2011). Chemometrics with R: Multivariate data analysis in the natural sciences and life sciences (pp. 155–165). New York: Springer.
Weljie, A. M., Bondareva, A., Zang, P., & Jirik, F. R. (2011). \(^1\)H-NMR metabolomics identification of markers of hypoxia-induced metabolic shifts in a breast cancer model system. Journal of Biomolecular NMR, 49(3–4), 185–193.
Wiklund, S., Johansson, E., Sjostrom, L., Mellerowicz, E., Edlund, U., Shockcor, J. P., et al. (2008). Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical Chemistry, 80(1), 115–122.
Wold, H. (1975). Path models with latent variables: The NIPALS approach (pp. 307–357). New York: Academic Press.
Wold, S., Trygg, J., Berglund, A., & Antti, H. (2002). Some recent developments in PLS modeling. Chemometrics and Intelligent Laboratory Systems, 58(2), 131–150.
Wold, S., Sjostrom, M., & Eriksson, L. (2001). PLS-regression: A basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems, 58(2), 109–130.
Zou, H., & Hastie, T. (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 67(2), 301–320.
Acknowledgements
The authors thank the GIGA-Cancer laboratory and Eli Lilly and Company for providing the data used in this paper. Support from the IAP Research Network P7/06 of the Belgian State (Belgian Science Policy) is also gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Ethical approval
This study analyzes collected data which involved human participants. For the q-PCR data set, the study was approved by our local Ethics Committee (CHR Citadelle, Liège, number B412201215082-1267) and all patients gave their informed consent.
Rights and permissions
About this article
Cite this article
Féraud, B., Munaut, C., Martin, M. et al. Combining strong sparsity and competitive predictive power with the L-sOPLS approach for biomarker discovery in metabolomics. Metabolomics 13, 130 (2017). https://doi.org/10.1007/s11306-017-1275-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1275-y