Abstract
Introduction
Pre-analytical processing significantly affects tissue metabolomes. Since most frozen kidney samples are stored after embedding, standardization of cryoprotective medium removal before metabolomics is essential.
Objectives
We used rodent and human kidney samples to develop an easy and robust pre-analytical procedure compatible with 1H-nuclear magnetic resonance (NMR)-based metabolomics.
Methods
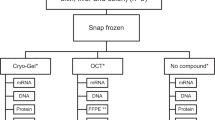
In mice, renal ischemia was induced for 30 min, followed by 48-h reperfusion (I/R, n = 6). Right kidneys were transversally cut in two fragments, and snap-frozen in liquid nitrogen (LN2) or in Optimal Cutting Temperature ® (OCT) fixative. In man, double kidney biopsies were simultaneously obtained before transplantation (n = 15), and snap-frozen in LN2 or OCT.
Results
1H-NMR spectrum of pure OCT highlighted two major peaks, i.e. from 3.4 to 4.2 ppm (47.2%) and from 1.2 to 2.2 ppm (42.5%). 1H-NMR spectra of mouse OCT kidneys were biased at 3.7. By contrast, 1H-NMR analyses of mouse OCT kidneys iteratively rinsed in saline significantly discriminated sham versus I/R groups, with Q² at 0.695 (to be compared with Q² at 0.866 for LN2 sham vs. I/R kidneys). Discriminant metabolites were analogous in both OCT and LN2 kidneys, with a correlation coefficient of 0.83. In man, iteratively rinsing OCT kidneys in saline eliminated the spectral 3.7-peak, thereby making metabolomes of OCT kidneys interpretable and similar to LN2 samples, with a correlation coefficient of 0.73.
Conclusion
NMR metabolomics using OCT-frozen kidney samples is valuable in mouse and man, following standardized OCT removal. This may help use residual biobanked human tissues to better understand renal pathophysiology.




Similar content being viewed by others
References
Atzler, D., Schwedhelm, E., & Zeller, T. (2014). Integrated genomics and metabolomics in nephrology. Nephrology, Dialysis, Transplantation, 29, 1467–1474.
Beckonert, O., Coen, M., Keun, H. C., Wang, Y., Ebbels, T. M., Holmes, E. et al. (2010). High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nature Protocols, 5, 1019–1032.
Cheung, C. C., Martin, B. R., & Asa, S. L. (2013). Defining diagnostic tissue in the era of personalized medicine. CMAJ, 185, 135–139.
Cisek, K., Krochmal, M., Klein, J., & Mischak, H. (2016). The application of multi-omics and systems biology to identify therapeutic targets in chronic kidney disease. Nephrology Dialysis Transplantation, 31, 2003–2011.
Frederich, M., Pirotte, B., Fillet, M., & de Tullio, P. (2016). Metabolomics as a challenging approach for medicinal chemistry and personalized medicine. Journal of Medicinal Chemistry, 59, 8649–8666.
Jouret, F., Leenders, J., Poma, L., Defraigne, J. O., Krzesinski, J. M., & de Tullio, P. (2016). Nuclear magnetic resonance metabolomic profiling of mouse kidney, urine and serum following renal ischemia/reperfusion injury. PLoS ONE, 11, e0163021.
Klein, J., Bascands, J. L., Mischak, H., & Schanstra, J. P. (2016). The role of urinary peptidomics in kidney disease research. Kidney International, 89, 539–545.
Martin, J. C., Maillot, M., Mazerolles, G., Verdu, A., Lyan, B., Migne, C. et al. (2015). Can we trust untargeted metabolomics? Results of the metabo-ring initiative, a large-scale, multi-instrument inter-laboratory study. Metabolomics, 11, 807–821.
Matheus, N., Hansen, S., Rozet, E., Peixoto, P., Maquoi, E., Lambert, V. et al. (2014). An easy, convenient cell and tissue extraction protocol for nuclear magnetic resonance metabolomics. Phytochemical Analysis: PCA, 25, 342–349.
Nicholson, J. K., Holmes, E., Kinross, J. M., Darzi, A. W., Takats, Z., & Lindon, J. C. (2012). Metabolic phenotyping in clinical and surgical environments. Nature, 491, 384–392.
Posada-Ayala, M., Zubiri, I., Martin-Lorenzo, M., Sanz-Maroto, A., Molero, D., Gonzalez-Calero, L. et al. (2014). Identification of a urine metabolomic signature in patients with advanced-stage chronic kidney disease. Kidney International, 85, 103–111.
Rhee, E. P. (2015). Metabolomics and renal disease. Current Opinion in Nephrology and Hypertension, 24, 371–379.
Righi, V., Schenetti, L., Maiorana, A., Libertini, E., Bettelli, S., Bonetti, L. R. et al. (2015). Assessment of freezing effects and diagnostic potential of BioBank healthy and neoplastic breast tissues through HR-MAS NMR spectroscopy. Metabolomics, 11, 487–498.
Serkova, N., Fuller, T. F., Klawitter, J., Freise, C. E., & Niemann, C. U. (2005). H-NMR-based metabolic signatures of mild and severe ischemia/reperfusion injury in rat kidney transplants. Kidney International, 67, 1142–1151.
Sieber, M., Hoffmann, D., Adler, M., Vaidya, V. S., Clement, M., Bonventre, J. V. et al. (2009). Comparative analysis of novel noninvasive renal biomarkers and metabonomic changes in a rat model of gentamicin nephrotoxicity. Toxicological Sciences, 109, 336–349.
Vivot, K., Benahmed, M. A., Seyfritz, E., Bietiger, W., Elbayed, K., Ruhland, E. et al. (2016). A metabolomic approach (1 H HRMAS NMR Spectroscopy) supported by histology to study early post-transplantation responses in islet-transplanted livers. International Journal of Biological Sciences, 12, 1168–1180.
Vrana, M., Goodling, A., Afkarian, M., & Prasad, B. (2016). An optimized method for protein extraction from OCT-embedded human kidney tissue for protein quantification by LC-MS/MS proteomics. Drug Metabolism and Disposition, 44, 1692–1696.
Weekers, L., de Tullio, P., Bovy, C., Poma, L., Maree, R., Bonvoisin, C. et al. (2015). Activation of the calcium-sensing receptor before renal ischemia/reperfusion exacerbates kidney injury. American Journal of Translational Research, 7, 128–138.
Weiss, R. H., & Kim, K. (2012). Metabolomics in the study of kidney diseases. Nature Reviews Nephrology, 8, 22–33.
Williams, W. W., Taheri, D., Tolkoff-Rubin, N., & Colvin, R. B. (2012). Clinical role of the renal transplant biopsy. Nature Reviews Nephrology, 8, 110–121.
Zhang, A., Sun, H., Qiu, S., & Wang, X. (2014). Metabolomics insights into pathophysiological mechanisms of nephrology. International Urology and Nephrology, 46, 1025–1030.
Zhang, W., Sakashita, S., Taylor, P., Tsao, M. S., & Moran, M. F. (2015). Comprehensive proteome analysis of fresh frozen and optimal cutting temperature (OCT) embedded primary non-small cell lung carcinoma by LC-MS/MS. Methods, 81, 50–55.
Acknowledgements
FJ is a Fellow of the Fonds National de la Recherche Scientifique (FNRS), and received support from the University of Liège (Fonds Spéciaux à la Recherche, Fonds Léon Fredericq) and the FNRS (Research Credit #3309). PDT is Senior Research Associate of the Fonds National de la Recherche Scientifique (FNRS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose.
Ethical approval
Our experimental mouse model was approved by the Institutional Animal Care and Use Committee of the University of Liège in Liège, Belgium (approval number: #1335). Our protocol using human kidney samples was approved by the Ethics Committee of the Cliniques Universitaires Saint-Luc (Université catholique de Louvain) in Brussels, Belgium (approval number; #2016/08MAR/086).
Rights and permissions
About this article
Cite this article
Leenders, J., Buemi, A., Mourad, M. et al. Nuclear magnetic resonance-based metabolomics of OCT-embedded frozen kidney samples in mouse and man following standardized pre-analytics. Metabolomics 13, 94 (2017). https://doi.org/10.1007/s11306-017-1232-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1232-9