Abstract
Introduction
Metabolomics investigates biochemical processes directly, potentially complementing transcriptomics and proteomics in providing insight into treatment outcome.
Objectives
This study aimed to use magnetic resonance (MR) spectroscopy on breast tumor tissue to explore the effect of neoadjuvant therapy on metabolic profiles, determine metabolic effects of the antiangiogenic drug bevacizumab, and investigate metabolic differences between responders and non-responders.
Methods
Breast tumors from 122 patients were profiled using high resolution magic angle spinning MR spectroscopy. All patients received neoadjuvant chemotherapy, and were randomized to receive bevacizumab or not. Tumors were biopsied prior, during, and after treatment.
Results
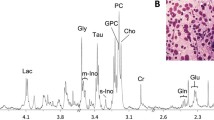
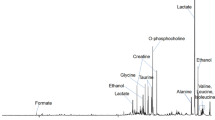
Principal component analysis showed clear metabolic changes indicating a decline in glucose consumption and a transition to normal breast adipose tissue as an effect of chemotherapy. Partial least squares-discriminant analysis revealed metabolic differences between pathological minimal residual disease patients and pathological non-responders after treatment (accuracy of 77%, p < 0.001), but not before or during treatment. Lower glucose and higher lactate was observed in patients exhibiting a good response (≥90% tumor reduction) compared to those with no response (≤10% tumor reduction) before treatment, while the opposite was observed after treatment. Bevacizumab-receiving and chemotherapy-only patients could not be discriminated at any time point. Linear mixed-effects models revealed a significant interaction between time and bevacizumab for glutathione, indicating higher levels of this antioxidant in chemotherapy-only patients than in bevacizumab receivers after treatment.
Conclusion
MR spectroscopy showed potential in detecting metabolic response to treatment and complementing other molecular assays for the elucidation of underlying mechanisms affecting pathological response.




Similar content being viewed by others
Abbreviations
- Ala:
-
Alanine
- Asc:
-
Ascorbate
- ATP:
-
Adenosine triphosphate
- Cho:
-
Choline
- Cr:
-
Creatine
- CV:
-
Cross validation
- ER:
-
Estrogen receptor
- FDR:
-
False discovery rate
- FEC:
-
5-Fluorouracil-epirubicin-cyclophosphamide
- Glc:
-
Glucose
- Glu:
-
Glutamate
- Gly:
-
Glycine
- GPC:
-
Glycerophosphocholine
- GR:
-
Good response
- GSH:
-
Glutathione
- HER2:
-
Human epidermal growth factor receptor 2
- HIF:
-
Hypoxia-inducible factor
- HR MAS MRS:
-
High resolution magic angle spinning magnetic resonance spectroscopy
- IL-8:
-
Interleukin 8
- IR:
-
Intermediate response
- Lac:
-
Lactate
- LMM:
-
Linear mixed-effects model
- LV:
-
Latent variable
- MICE:
-
Multiple imputation by chained equations
- MR:
-
Magnetic resonance
- MRI:
-
Magnetic resonance imaging
- NR:
-
No response
- OPLS:
-
Orthogonal partial least squares
- OS:
-
Overall survival
- PAM50:
-
Prediction analysis of microarrays 50
- PC:
-
Principal component
- PCA:
-
Principal component analysis
- PCh:
-
Phosphocholine
- pCR:
-
Pathological complete response
- PFS:
-
Progression-free survival
- PLS-DA:
-
Partial least squares-discriminant analysis
- pMRD:
-
Pathological minimal residual disease
- pNR:
-
Pathological non-responder
- PQN:
-
Probabilistic quotient normalization
- q–q:
-
Quantile–quantile
- RMSE:
-
Root mean square error
- ROS:
-
Reactive oxygen species
- R2 :
-
Coefficient of determination
- SDH:
-
Succinate dehydrogenase
- Succ:
-
Succinate
- Tau:
-
Taurine
- TCA:
-
Tricarboxylic acid
- TP:
-
Time point
- VEGF:
-
Vascular endothelial growth factor
References
Aboagye, E. O., & Bhujwalla, Z. M. (1999). Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Research, 59(1), 80–84.
American Cancer Society (2011). Global cancer facts & figures (2nd ed.). Atlanta: American Cancer Society.
Bathen, T. F., Sitter, B., Sjøbakk, T. E., Tessem, M.-B., & Gribbestad, I. S. (2010). Magnetic resonance metabolomics of intact tissue: A biotechnological tool in cancer diagnostics and treatment evaluation. Cancer Research, 70(17), 6692–6696. doi:10.1158/0008-5472.can-10-0437.
Bear, H. D., Tang, G., Rastogi, P., Geyer, C. E. Jr., Liu, Q., Robidoux, A., et al. (2015). Neoadjuvant plus adjuvant bevacizumab in early breast cancer (NSABP B-40 [NRG Oncology]): Secondary outcomes of a phase 3, randomised controlled trial. The Lancet Oncology, 16(9), 1037–1048. doi:10.1016/S1470-2045(15)00041-8.
Borgan, E., Sitter, B., Lingjærde, O. C., Johnsen, H., Lundgren, S., Bathen, T. F., et al. (2010). Merging transcriptomics and metabolomics—advances in breast cancer profiling. BMC Cancer, 10. doi:10.1186/1471-2407-10-628.
Cao, M. D., Giskeødegård, G. F., Bathen, T. F., Sitter, B., Bofin, A., Lonning, P. E., et al. (2012). Prognostic value of metabolic response in breast cancer patients receiving neoadjuvant chemotherapy. BMC Cancer, 12. doi:10.1186/1471-2407-12-39.
Cao, M. D., Sitter, B., Bathen, T. F., Bofin, A., Lonning, P. E., Lundgren, S., et al. (2011). Predicting long-term survival and treatment response in breast cancer patients receiving neoadjuvant chemotherapy by MR metabolic profiling. NMR in Biomedicine, 25. doi:10.1002/nbm.1762.
Dieterle, F., Ross, A., Schlotterbeck, G., & Senn, H. (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1 H NMR metabonomics. Analytical Chemistry, 78(13), 4281–4290. doi:10.1021/ac051632c.
Eigenvector Research. (2013). Orthogonalizepls. Retrieved August 2015, from http://wiki.eigenvector.com/index.php?title=Orthogonalizepls.
Fack, F., Espedal, H., Keunen, O., Golebiewska, A., Obad, N., Harter, P., et al. (2015). Bevacizumab treatment induces metabolic adaptation toward anaerobic metabolism in glioblastomas. Acta Neuropathologica, 129(1), 115–131. doi:10.1007/s00401-014-1352-5.
Ferlay, J., Soerjomataram, I., Ervik, M., Dikshit, R., Eser, S., Mathers, C., et al. (2013). GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC Cancer Base No. 11. Retrieved July 21, 2015, from http://globocan.iarc.fr.
Feron, O. (2009). Pyruvate into lactate and back: From the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiotherapy and Oncology, 92(3), 329–333. doi:10.1016/j.radonc.2009.06.025.
Ferrara, N., Hillan, K. J., Gerber, H.-P., & Novotny, W. (2004). Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nature Reviews Drug Discovery, 3(5), 391–400. doi:10.1038/nrd1381.
Franco, R., & Cidlowski, J. A. (2009). Apoptosis and glutathione: Beyond an antioxidant. Cell Death & Differentiation, 16(10), 1303–1314.
Gibellini, F., & Smith, T. K. (2010). The Kennedy pathway—de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life, 62(6), 414–428. doi:10.1002/iub.337.
Giordano, S. H., Buzdar, A. U., Smith, T. L., Kau, S.-W., Yang, Y., & Hortobagyi, G. N. (2004). Is breast cancer survival improving? Cancer, 100(1), 44–52. doi:10.1002/cncr.11859.
Giskeødegård, G. F., Cao, M. D., & Bathen, T. F. (2015). High-resolution magic-angle-spinning NMR spectroscopy of intact tissue. In J. T. Bjerrum (Ed.), Metabonomics (Vol. 1277, pp. 37–50, Methods in molecular biology). New York: Springer New York.
Giskeødegård, G. F., Grinde, M. T., Sitter, B., Axelson, D. E., Lundgren, S., Fjøsne, H. E., et al. (2010). Multivariate modeling and prediction of breast cancer prognostic factors using MR metabolomics. Journal of Proteome Research, 9(2), 972–979. doi:10.1021/pr9008783.
Giskeødegård, G. F., Lundgren, S., Sitter, B., Fjøsne, H. E., Postma, G., Buydens, L. M. C., et al. (2012). Lactate and glycine—potential MR biomarkers of prognosis in estrogen receptor-positive breast cancers. NMR in Biomedicine, 25(11), 1271–1279. doi:10.1002/nbm.2798.
Glunde, K., Bhujwalla, Z. M., & Ronen, S. M. (2011). Choline metabolism in malignant transformation. Nature Reviews Cancer, 11(12), 835–848. doi:10.1038/nrc3162.
Grinde, M. T., Skrbo, N., Moestue, S. A., Rødland, E. A., Borgan, E., Kristian, A., et al. (2014). Interplay of choline metabolites and genes in patient-derived breast cancer xenografts. Breast Cancer Research, 16, R5. doi:10.1186/bcr3597.
Gyanchandani, R., Sano, D., Ortega Alves, M. V., Klein, J. D., Knapick, B. A., Oh, S., et al. (2013). Interleukin-8 as a modulator of response to bevacizumab in preclinical models of head and neck squamous cell carcinoma. Oral Oncology, 49(8), 761–770. doi:10.1016/j.oraloncology.2013.03.452.
Hanahan, D., & Weinberg, R. A. (2000). The hallmarks of cancer. Cell, 100(1), 57–70. doi:10.1016/S0092-8674(00)81683-9.
Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell, 144(5), 646–674. doi:10.1016/j.cell.2011.02.013.
Haukaas, T. H., Euceda, L. R., Giskeødegård, G. F., Lamichhane, S., Krohn, M., Jernström, S., et al. (2016). Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer & Metabolism, 4(1), 12. doi:10.1186/s40170-016-0152-x.
Hensley, C. T., Wasti, A. T., & DeBerardinis, R. J. (2013). Glutamine and cancer: Cell biology, physiology, and clinical opportunities. The Journal of Clinical Investigation, 123(9), 3678–3684. doi:10.1172/JCI69600.
Huang, C., & Freter, C. (2015). Lipid metabolism, apoptosis and cancer therapy. International Journal of Molecular Sciences, 16(1), 924.
King, A., Selak, M. A., & Gottlieb, E. (2006). Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene, 25(34), 4675–4682.
Malmgren, J. A., Parikh, J., Atwood, M. K., & Kaplan, H. G. (2014). Improved prognosis of women aged 75 and older with mammography-detected breast cancer. Radiology, 273(3), 686–694. doi:10.1148/radiol.14140209.
Miller, E., Lee, H., Lulla, A., Hernandez, L., Gokare, P., & Lim, B. (2014). Current treatment of early breast cancer: Adjuvant and neoadjuvant therapy. F1000Research, 3, 198.
Miller, K., Wang, M., Gralow, J., Dickler, M., Cobleigh, M., Perez, E. A., et al. (2007). Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. New England Journal of Medicine, 357(26), 2666–2676. doi:10.1056/NEJMoa072113.
Moestue, S., Sitter, B., Bathen, T. F., Tessem, M.-B., & Gribbestad, I. S. (2011). HR MAS MR spectroscopy in metabolic characterization of cancer. Current Topics in Medicinal Chemistry, 11(1), 2–26.
Moestue, S. A., Giskeødegård, G. F., Cao, M. D., Bathen, T. F., & Gribbestad, I. S. (2012). Glycerophosphocholine (GPC) is a poorly understood biomarker in breast cancer. Proceedings of the National Academy of Sciences of the United States of America, 109(38), E2506. doi:10.1073/pnas.1208226109.
Ng, C. K., Pemberton, H. N., & Reis-Filho, J. S. (2012). Breast cancer intratumor genetic heterogeneity: Causes and implications. Expert Review of Anticancer Therapy, 12(8), 1021–1032. doi:10.1586/era.12.85.
Parker, J. S., Mullins, M., Cheang, M. C. U., Leung, S., Voduc, D., Vickery, T., et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of Clinical Oncology, 27(8), 1160–1167. doi:10.1200/jco.2008.18.1370.
Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., et al. (2000). Molecular portraits of human breast tumours. Nature, 406(6797), 747–752. doi:10.1038/35021093.
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D., & R Core Team (2014) (2014). nlme: Linear and nonlinear mixed effects models. R package version 3.1–117. http://CRAN.R-project.org/package=nlme.
Pinheiro, J. C., & Bates, D. M. (2000). Linear mixed-effects models: Basic concepts and examples. In Mixed-effects models in S and S-PLUS (pp. 3–56, Statistics and computing). New York: Springer New York.
R Core Team (2014). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. http://www.R-project.org/.
Rubin, D. B. (1987). Mulitiple imputation for nonresponse in surveys. New York: Wiley.
Saraswathy, S., Crawford, F., Lamborn, K., Pirzkall, A., Chang, S., Cha, S., et al. (2009). Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. Journal of Neuro-Oncology, 91(1), 69–81. doi:10.1007/s11060-008-9685-3.
Selak, M. A., Armour, S. M., MacKenzie, E. D., Boulahbel, H., Watson, D. G., Mansfield, K. D., et al. (2005). Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell, 7(1), 77–85. doi:10.1016/j.ccr.2004.11.022.
Semenza, G. L. (2008). Tumor metabolism: Cancer cells give and take lactate. The Journal of Clinical Investigation, 118(12), 3835–3837. doi:10.1172/JCI37373.
Sitter, B., Sonnewald, U., Spraul, M., Fjösne, H. E., & Gribbestad, I. S. (2002). High-resolution magic angle spinning MRS of breast cancer tissue. NMR in Biomedicine, 15(5), 327–337. doi:10.1002/nbm.775.
Sørlie, T., Tibshirani, R., Parker, J., Hastie, T., Marron, J. S., Nobel, A., et al. (2003). Repeated observation of breast tumor subtypes in independent gene expression data sets. Proceedings of the National Academy of Sciences of the United States of America, 100(14), 8418–8423. doi:10.1073/pnas.0932692100.
van Buuren, S., & Groothuis-Oudshoorn, K. (2011). mice: Multivariate imputation by chained equations in R. Journal of Statistical Software, 45(3), 1–67.
van der Hage, J. A., van de Velde, C. J. H., Julien, J.-P., Tubiana-Hulin, M., Vandervelden, C., Duchateau, L., et al. (2001). Preoperative chemotherapy in primary operable breast cancer: Results from the European Organization for research and treatment of cancer trial 10902. Journal of Clinical Oncology, 19(22), 4224–4237.
Vander Heiden, M. G., Cantley, L. C., & Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science, 324(5930), 1029–1033. doi:10.1126/science.1160809.
Végran, F., Boidot, R., Michiels, C., Sonveaux, P., & Feron, O. (2011). Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Research, 71(7), 2550–2560. doi:10.1158/0008-5472.can-10-2828.
Walenta, S., & Mueller-Klieser, W. F. (2004). Lactate: Mirror and motor of tumor malignancy. Seminars in Radiation Oncology, 14(3), 267–274. doi:10.1016/j.semradonc.2004.04.004.
Walenta, S., Wetterling, M., Lehrke, M., Schwickert, G., Sundfør, K., Rofstad, E. K., et al. (2000). High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Research, 60(4), 916–921.
Westerhuis, J., Hoefsloot, H. J., Smit, S., Vis, D., Smilde, A., van Velzen, E. J., et al. (2008). Assessment of PLSDA cross validation. Metabolomics, 4(1), 81–89. doi:10.1007/s11306-007-0099-6.
Wold, S., Esbensen, K., & Geladi, P. (1987). Principal component analysis. Chemometrics and Intelligent Laboratory Systems, 2, 37–52.
Wold, S., Sjöström, M., & Eriksson, L. (2001). PLS-regression: A basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems, 58(2), 109–130. doi:10.1016/S0169-7439(01)00155-1.
Xie, J., Wu, H., Dai, C., Pan, Q., Ding, Z., Hu, D., et al. (2014). Beyond Warburg effect—dual metabolic nature of cancer cells. Scientific Reports, 4. doi:10.1038/srep04927.
Acknowledgements
The authors would like to thank Øyvind Salvesen for useful discussions regarding linear mixed-effects models and Santosh Lamichhane for technical support during HR MAS MRS acquisition. The HR MAS MRS analysis was performed at the MR Core Facility, Norwegian University of Science and Technology (NTNU), which is funded by the Faculty of Medicine and Health Sciences at NTNU and the Central Norway Regional Health Authority. The study was funded in part by generous grants from: (1) The Research Council of Norway, Imaging the breast cancer metabolome, Project no 221879, (2) The Pink Ribbon Movement and Norwegian Breast Cancer Society, (3) K. G. Jebsen Center for Breast Cancer Research, (4) Roche Norway, (5) Sanofi-Aventis Norway. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The NeoAva study was co-sponsored by Roche Norway and Sanofi-Aventis Norway. Oslo University Hospital is the main sponsor for the NeoAva study.
Authors' contributions
LRE, THH, GFG, RV, JE, LSP, GP, LMCB, ALBD, OE, and TFB participated in the design of the study. ALBD, OE, and TFB conceived the study. LRE, THH, GFG, RV, JE, GP, LMCB, ALBD, OE, and TFB interpreted the data. LRE performed the HR MAS MRS acquisition, statistical analysis, and drafted the manuscript. LSP, SL, EB, OG, ALBD, OE, and TFB participated in acquisition of the data. All authors have read and helped to revise the manuscript. The final manuscript is approved by all the authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
The study was approved for all centers involved by the Regional Ethics Committee (Approval number S-08354a) and the Norwegian Medical Agency.
Research involving human and animal rights
All procedures performed in studies involving human participants were in accordance with the Declaration of Helsinki, International Conference on Harmony/Good Clinical practice.
Informed consent
Informed written consent was obtained from all patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Euceda, L.R., Haukaas, T.H., Giskeødegård, G.F. et al. Evaluation of metabolomic changes during neoadjuvant chemotherapy combined with bevacizumab in breast cancer using MR spectroscopy. Metabolomics 13, 37 (2017). https://doi.org/10.1007/s11306-017-1168-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-017-1168-0