Abstract
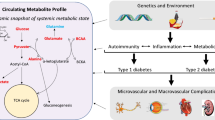
Biomarker studies for metabolic disorders like diabetes mellitus (DM) are an important approach towards a better understanding of the underlying pathophysiological mechanisms of diseases (Roberts and Gerszten in Cell Metab 18:43–50, 2013; Wilson et al. in Proteome Res 4:591–598, 2005). Furthermore, screening of potential metabolic biomarkers opens the opportunity of early diagnosis as well as therapy and drug monitoring of metabolic disorders (Rhee et al. in J Clin Invest 10:1–10, 2011; Wang et al. in Nat Med 17:448–458, 2011; Wenk in Nat Rev Drug Discov 4:594–610, 2005). The aim of the present study was to develop methods for the quantitative determination of 74 potential metabolite biomarkers for DM and diabetic nephropathy (DN) in serum. Several studies have shown that the concentrations of many polar metabolites like amino or organic acids are changed in subjects suffering from diabetes (Wang et al. in Nat Med 17:448–458, 2011; Yuan et al. in J Chromatogr B 813:53–58, 2007). Analyzing polar analytes presents a challenge in liquid chromatography (LC) coupled with ESI–MS/MS (Gika et al. in J Sep Sci 31:1598–1608, 2008; Spagou et al. in J Sep Sci 33:716–727, 2010). Considering those reasons we decided to develop a specific HILIC–ESI–QqQ–MS/MS-method for quantitative determination of these polar metabolites. A subsequent method validation was carried out for both HILIC and RP chromatography with respect to the guidelines of the Food and Drug Administration (FDA in Food and Drug Administration: Guidance for industry, bioanalytical method validation, 2001). The HILIC and RP LC–MS methods were successfully validated. Furthermore, the HILIC method presented here was applied to serum samples of GIPRdn transgenic mice, a diabetic strain developing DN, and non transgenic littermate controls. Significant, diabetes-associated changes were observed for the concentrations of 21 out of 62 metabolites. The new methods described here accurately quantify 74 metabolites known to be regulated in diabetes, allowing for direct comparison between studies and laboratories. Thus, these methods may be highly adoptable in clinical research, providing a starting point for early diagnosis and metabolic screening.



Similar content being viewed by others
References
Altmaier, E., Ramsay, S. L., Graber, A., Mewes, H.-W., Weinberger, K. M., & Suhre, K. (2008). Bioinformatics analysis of targeted metabolomics—Uncovering old and new tales of diabetic mice under medication. Endocrinology, 149, 3478–3489.
Bao, Y., et al. (2009). Metabonomic variations in the drug-treated Type 2 diabetes mellitus patients and healthy volunteers. Journal of Proteome Research, 8, 1623–1630.
Bonnefond, A., & Froguel, P. (2015). Rare and common genetic events in type 2 diabetes: What should biologists know? Cell Metabolism, 21, 357–368.
Chen, P., & Liu, J. (2007). Metabonomics and diabetes mellitus. Advances in Therapy, 24, 1036–1045.
Ciapaite, J., Bakker, S. J. L., Heine, R. J., Krab, K., & Westerhoff, H. V. (2007). A systems biology perspective on obesity and Type 2 diabetes. In V. Saks (Ed.), Molecular system bioenergetics: Energy for life (pp. 571–592). Weinheim: Wiley.
Connor, S. C., Hansen, M. K., Corner, A., Smith, R. F., & Ryan, T. E. (2010). Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Molecular BioSystems, 6, 909–921.
Demetz, E., et al. (2014). The arachidonic acid metabolome serves as a conserved regulator of cholesterol metabolism. Cell Metabolism, 20, 787–798.
FDA. (2001). Food and Drug Administration: Guidance for industry, bioanalytical method validation. Rockville: Department of Health and Human Services.
Fiehn, O., & Kind, T. (2007). Metabolite profiling in blood plasma. Methods in Molecular Biology, 358, 3–17.
Galazis, N., Iacovou, C., Haoula, Z., & Atiomo, W. (2012). Metabolomic biomarkers of impaired glucose tolerance and type 2 diabetes mellitus with a potential for risk stratification in women with polycystic ovary syndrome. European Journal of Obstetrics & Gynecology and Reproductive Biology, 160, 121–130.
Gika, H. G., Theodoridis, G. A., & Wilson, I. D. (2008). Hydrophilic interaction and reversed-phase ultraperformance liquid chromatography TOF-MS for metabonomic analysis of Zucker rat urine. Journal of Separation Science, 31, 1598–1608.
Godzien, J., et al. (2011). Metabolomic approach with LC-QTOF to study the effect of a nutraceutical treatment on urine of diabetic rats. Journal of Proteome Research, 10, 837–844.
Herbach, N., Goeke, B., Schneider, M., Hermanns, W., Wolf, E., & Wanke, R. (2005). Overexpression of a dominant negative GIP receptor in transgenic mice results in disturbed postnatal pancreatic islet and beta-cell development. Regulatory Peptides, 125, 103–117.
Herbach, N., et al. (2009). Diabetic kidney lesions of GIPRdn transgenic mice: podocyte hypertrophy and thickening of the GBM precede glomerular hypertrophy and glomerulosclerosis. American Journal of Physiology Renal Physiology, 296, 819–829.
Huang, Q., et al. (2011). Method for liver tissue metabolic profiling study and its application in type 2 diabetic rats based on ultra performance liquid chromatography-mass spectrometry. Journal of Chromatography B, 879, 961–967.
IDF. (2011). International diabetes federation annual report (pp. 1–36). Belgium: Brussel.
Ismail, A. A., & Gill, G. V. (1999). The epidemiology of Type 2 diabetes and its current measurement. Baillière’s Best Practice & Research Clinical Endocrinology & Metabolism, 13, 197–220.
John, C., et al. (2014). A liquid chromatography-tandem mass spectrometry-based method for the simultaneous determination of hydroxy sterols and bile acids. Journal of Chromatography A, 1371, 184–195.
Kanehisa, M. (2002). The KEGG database. Novartis Found Symp, 247, 91–101. discussion 101–103, 119–128, 244–252.
Magnusson, M., et al. (2015). Dimethylglycine deficiency and the development of diabetes mellitus. Diabetes, 64, 3010–3016.
Malecki, M. T. (2005). Genetics of type 2 diabetes mellitus. Diabetes Research and Clinical Practice, 68, S10–S21.
Ohlson, L.-O., et al. (1988). Risk factors for type 2 (non-insulin-dependent) diabetes mellitus. Thirteen and one-half years of follow-up of the participants in a study of Swedish men born in 1913. Diabetologia, 31, 798–805.
Patti, G. J., Yanes, O., & Siuzdak, G. (2012). Innovation: Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13, 263–269.
Preet, A., Karve, T. M., Rizk, N., & Cheema, K. (2012). Metabolomics: Approaches and applications to diabetes research. Journal of Diabetes & Metabolism, S6, 001.
Puri, P., et al. (2009). The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology, 50, 1827–1838.
Rhee, E. P., et al. (2011). Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. The Journal of Clinical Investigation, 10, 1–10.
Roberts, L. D., & Gerszten, R. E. (2013). Toward new biomarkers of cardiometabolic diseases. Cell Metabolism, 18, 43–50.
Spagou, K., Tsoukali, H., Raikos, N., Gika, H., Wilson, I. D., & Theodoridis, G. (2010). Hydrophilic interaction chromatography coupled to MS for metabonomic/metabolomic studies. Journal of Separation Science, 33, 716–727.
Tsutsui, H., et al. (2010). Practical analytical approach for the identification of biomarker candidates in prediabetic state based upon metabonomic study by ultraperformance liquid chromatography coupled to electrospray ionization time-of-flight mass spectrometry. Journal of Proteome Research, 9, 3912–3922.
Tuomilehto, J., et al. (2001). Prevention of Type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. The New England Journal of Medicine, 344, 1343–1350.
Wang, T. J., et al. (2011). Metabolite profiles and the risk of developing diabetes. Nature Medicine, 17, 448–453.
Wenk, M. R. (2005). The emerging field of lipidomics. Nature Reviews Drug Discovery, 4, 594–610.
Wilson, I. D., et al. (2005). High resolution “Ultra performance” liquid chromatography coupled to oa-TOF mass spectrometry as a tool for differential metabolic pathway profiling in functional genomic studies. Journal of Proteome Research, 4, 591–598.
Xu, J., Zhang, J., Dong, J., Cai, S., Yang, J. Y., & Chen, Z. (2009). Metabonomics studies of intact hepatic and renal cortical tissues from diabetic db/db mice using high-resolution magic-angle spinning 1H NMR spectroscopy. Analytical and Bioanalytical Chemistry, 393, 1657–1668.
Yang, J., et al. (2004). Discrimination of Type 2 diabetic patients from healthy controls by using metabonomics method based on their serum fatty acid profiles. Journal of Chromatography B, 813, 53–58.
Yuan, K., Kong, H., Guan, Y., Yang, J., & Xu, G. (2007). A GC-based metabonomics investigation of type 2 diabetes by organic acids metabolic profile. Journal of Chromatography B, 850, 236–240.
Zhao, T., et al. (2012). Intrarenal metabolomics reveals the association of local organic toxins with the progression of diabetic kidney disease. Journal of Pharmaceutical and Biomedical Analysis, 60, 32–43.
Zhu, C., Liang, Q. L., Hu, P., Wang, Y. M., & Luo, G. A. (2011). Phospholipidomic identification of potential plasma biomarkers associated with type 2 diabetes mellitus and diabetic nephropathy. Talanta, 85, 1711–1720.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Ernst Meiss, Philipp Werner, and Clara John have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Meiss, E., Werner, P., John, C. et al. Metabolite targeting: development of a comprehensive targeted metabolomics platform for the assessment of diabetes and its complications. Metabolomics 12, 52 (2016). https://doi.org/10.1007/s11306-016-0958-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-016-0958-0