Abstract
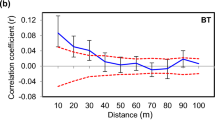
To achieve gene preservation in tree populations when planting seedlings to regenerate forests, specific and practical guidelines and criteria for seed collection are needed to ensure reliable coverage and effective capture of the current genetic variation. We examined the genetic variation of adult trees (524 trees) and seed pools (1618 seeds) collected from 70 mother trees in an old planted population of Pinus thunbergii in Japan using seven nuclear microsatellite markers. To consider a suitable seed collection strategy, we monitored changes in allelic diversity of seed pools with an increasing number of mother trees and examined the required number of mother trees. We found significantly higher allelic diversity statistics in seed pools of mother trees with smaller diameter at breast height. Spatial genetic autocorrelation analysis showed significantly positive kinship coefficients between both pairs of adult trees and seeds collected from mother trees for up to 100-m distance classes. Based on the rarefaction curve, seed pools obtained from approximately up to 30 mother trees could cover and saturate most of the genetic diversity and composition of the adult tree population and the potential overall seed pools, particularly for statistics that are less likely affected by rare alleles. Indications from the mother tree number, together with considerations for the size or spatial distribution of mother trees, are expected to contribute not only to guidelines for in situ genetic management of local planted populations, but also to the development of strategies for ex situ gene preservation of populations as genetic resources.



Similar content being viewed by others
Data availability
Location, size, and genotype data of adult tree population and seed pools are available via online as the supplementary materials.
References
Arista M, Talavera S (1997) Gender expression in Abies pinsapo Boiss., a Mediterranean fir. Ann Bot 79:337–342
Austerlitz F, Smouse PE (2001) Two-generation analysis of pollen flow across a landscape. II. Relation between ΦFT, pollen dispersal, and inter-female distance. Genet 157:851–857
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genet 144:2001–2014
Dow BD, Ashley MV (1996) Microsatellite analysis of seed dispersal and parentage of saplings in bur oak, Quercus macrocarpa. Mol Ecol 5:615–627
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
El-Kassaby YA (2003) Clonal-row vs. random seed orchard designs: mating pattern and seed yield of western hemlock (Tsuga heterophylla (Raf.) Sarg.). For Genet 10:121–127
Furukoshi T (1991) A gene bank project for preservation of forest tree genetic resources. In: Ohba K, Katsuta M (eds.) Forest tree breeding. Bun-eido Shuppan, Tokyo, pp 224–241.
Goto S, Shimatani K, Yoshimaru H, Takahashi Y (2006) Fat-tailed gene flow in the dioecious canopy tree species Fraxinus mandshurica var. japonica revealed by microsatellites. Mol Ecol 15:2985–2996
Goudet J (2002) FSTAT version 2.9.3, a program to estimate and test gene diversities and fixation indices [computer program]. Available from: http://www.unil.ch/izea/softwares/fstat.html.
Guichoux E, Lagache L, Wagne S, Chaumeil P, Leger P, Lepais O, Lepoittevi C, Malausa T, Revardel E, Salin F, Petit RJ (2011) Current trends in microsatellite genotyping. Mol Ecol Resour 11:591–611
Hale ML, Burg TM, Steeves TE (2012) Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS ONE 7:e45170
Hamrick JL, Godt MJW, Sherman-Broyles SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forest 6:95–124
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hoban S, Schlarbaum S (2014) Optimal sampling of seeds from plant populations for ex-situ conservation of genetic biodiversity, considering realistic population structure. Biol Cons 177:90–99
Hosius B, Leinemann L, Konnert M, Bergmann F (2006) Genetic aspects of forestry in the Central Europe. Eur J Forest Res 125:407–417
Irwin AJ, Hamrick JL, Godt MJW, Smouse PE (2003) Significance multiyear estimate of the effective pollen donor pool for Albizia julibrissin. Hered 90:187–194
Iwaizumi MG, Takahashi M, Watanabe A, Ubukata M (2010) Simultaneous evaluation of paternal and maternal immigrant gene flow and the implications for the overall genetic composition of Pinus densiflora dispersed seeds. J Hered 101:144–153
Iwaizumi MG, Takahashi M, Isoda K, Austerlitz F (2013) Consecutive five-year analysis of paternal and maternal gene flow and contributions of the gametic heterogeneities to overall genetic composition of Pinus densiflora dispersed seeds. Am J Bot 100:1896–1904
Iwaizumi MG, Miyata S, Hirao T, Tamura M, Watanabe A (2018) Historical seed use and transfer affects geographic specificity in genetic diversity and structure of old planted Pinus thunbergii populations. For Ecol Manag 408:211–219
Iwaizumi MG, Takahashi M, Yano K (2021) Temporal variation in regeneration events affecting population structure in different size- and life-stages contributes to overall genetic diversity of natural Zelkova serrata population. J for Res 26:32–42
Iwaizumi MG, Matsunaga K, Iki T, Yamanobe T, Hirao T, Watanabe A (2021) Geographical cline and inter-seaside difference in cone characteristics related to climatic conditions of old planted Pinus thunbergii populations throughout Japan. Plant Species Biol 36:218–229
Iwaizumi MG, Nasu J, Miyamoto N, Isoda K (2021) Genetic variation of seed pools in two mast years and a genetic preservation strategy for a population of the endemic conifer Abies veitchii var. Shikokiana. J Jpn for Soc 103:172–179 (in Japanese with English summary)
Iwaizumi MG, Ohtani M, Yano K, Nasu J, Miyamoto N, Takahashi M, Ubukata M (2022) Accurate paternal and maternal immigrant gene flow analysis over three mast years detects relative levels of gametic heterogeneity components in a natural population of Abies firma. J For Res 27:297–307
Iwasaki H, Uchiyama K, Kimura MK, Saito Y, Hakamata T, Ide Y (2019) Impact of a tree improvement program on the genetic diversity of sugi (Cryptomeria japonica D Don) plantations. For Ecol Manag 448:466–473
Kanda Y (2013) Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marr Transplant 48:452–458
Kohyama T (1982) Studies on the Abies population of Mt. Shimagare II. Reproductive and life history traits. Bot Mag Tokyo 95:167–181
Kim Y-M, Hong K-N, Park Y-J, Hong Y-P, Park J-I (2015) Estimating the parameters of pollen flow and mating system in Pinus densiflora population in Buan, South Korea, using microsatellite markers. Korean J Plant Res 28:101–110
Li G, Yang M, Zhou K, Zhang L, Tian L, Lv S, Jin Y, Qian W, Xiong H, Lin R, Fu Y, Hou X (2015) Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol 25:1136–1145
Loicelle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinails (Rubiaceae). Am J Bot 82:1420–1425
Mamiya Y (1988) History of pine wilt disease in Japan. J Nematol 20:219–226
Marshall DR, Brown AHD (1975) The charge-state model of protein polymorphism in natural populations. J Mol Evol 6:149–163
Miyamoto N, Fernandez-Manjarres JF, Morand-prieur M-E, Bertolino P, Frascaria-Lacoste N (2008) What sampling is needed for reliable estimations of genetic diversity in Fraxinus excelsior L. (Oleaceae)? Ann For Sci 65:403
Miyata M, Ubukata M (1994) Genetic variations of allozymes in natural stands of Japanese black pine. J Jpn for Soc 76:445–455 (In Japanese with English sumamary)
Morgante M, Vendramin GG, Rossi P (1991) Effects of stand density on outcrossing rate in two Norway spruce (Picea abies) populations. Can J Bot 69:2704–2708
Mukasyaf AA, Matsunaga K, Tamura M, Iki T, Watanabe A, Iwaizumi MG (2021) Reforestation or genetic disturbance: a case study of Pinus thunbergii in the Iki-no-Matsubara coastal forest (Japan). Forests 12:72
Nakanishi A, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S (2005) Interannual genetic heterogeneity of pollen pools accepted by Quercus salicina individuals. Mol Ecol 14:4469–4478
Nakayama M, Kobayashi Y (1981) Pinus Linn. In Asakawa S, Katsuta M, Yokoyama T (eds.) Seeds of woody plants in Japan: Gymnospermae. Japan Forest Tree Breeding Association, Tokyo, pp. 65–76 (in Japanese).
Nathan R, Katul CG, Horn HS, Thomas SM, Oren R, Avissar R, Pacala SW, Levin SA (2002) Mechanisms of long-distance dispersal of seeds by wind. Nat 418:409–413
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Petit RJ, El Mousadik A, Pons O (1998) Identifying population for conservation on the basis of genetic markers. Conserv Biol 12:844–855
R Development Core Team (2011) R: a language and environment for statistical computing. http://www.R-project.org.
Ritland K (2002) Extensions of models for the estimation of mating systems using n independent loci. Hered 88:221–228
Robledo-Arnuncio JJ, Smouse PE, Gil L, Alia R (2004) Pollen movement under alternative silvicultural practices in native populations of Scots pine (Pinus sylvestris L.) in central Spain. For Ecol Manag 197:243–253
Robledo-Arnuncio JJ, Austerlitz F, Smouse PE (2007) POLDISP: a software package for indirect estimation of contemporary pollen dispersal. Mol Ecol Notes 7:763–766
Shiraishi S, Watanabe A (1995) Identification of chloroplast genome between Pinus densiflora Sieb. et Zucc. and Pinus thunbergii Parl. based on the polymorphism in rbcL gene. J Jpn for Soc 77:429–436 (in Japanese with English summary)
Sjogren P, Wyoni PI (1994) Conservation genetics and detection of rare alleles in finite populations. Conserv Biol 8:267–270
Smouse PE, Dyer RJ, Westfall RD, Sork VL (2001) Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evol 55:260–271
Troupin D, Nathan R, Vendramin GG (2006) Analysis of spatial genetic structure in an expanding Pinus halepensis population reveals development of fine-scale genetic clustering over time. Mol Ecol 15:3617–3630
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Volk GM, Richards CM, Reilley AA, Henk AD, Forsline PL, Aldwinckle HS (2005) Ex-situ conservation of vegetatively propagated species: development of seed-based core collection for Malus sieversii. J Am Soc Hort Sci 130:203–210
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgements
We thank M. Kubota, H. Yamada, H. Miyashita, M. Miura, Y. Kawai-Munehara, and other members of the Forest Tree Breeding Center and Kyushu University, for kind collaboration in field surveys and cone samplings and also the continued advices and encouragements during the present study. We express our special thanks to M. Shimoyama for her assistance in the DNA analyzes. The present study was funded by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grant-in-Aid for Scientific Research, No. 17K07853).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by J. Beaulieu
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iwaizumi, M.G., Mukasyaf, A.A., Tamaki, I. et al. Genetic diversity and structure of seed pools in an old planted Pinus thunbergii population and seed collection strategy for gene preservation. Tree Genetics & Genomes 19, 9 (2023). https://doi.org/10.1007/s11295-022-01584-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-022-01584-5