Abstract
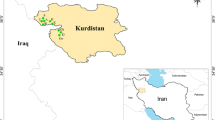
Quercus infectoria, commonly known as gall oak, is a small shrub found in Iran. Unfortunately, it is subjected to genetic erosion, and so, its conservation and evaluation are desirable. Thus, in the current research, 16 microsatellite primer pairs (seven nuclear simple sequence repeats (nSSRs) and nine chloroplast simple sequence repeats (cpSSRs)) were used in an attempt to assess the genetic diversity of 121 individuals of Q. infectoria belonging to 11 populations from three provinces in northern Zagros forests of Iran. In total, 69 alleles of nSSR and 18 alleles of cpSSR were detected among the individuals. The results of the overall analysis of molecular variance based on nSSRs indicated that 89.00% of the variation was due to differences within populations and 11.00% occurred among populations, while according to cpSSRs, 94.00% of the variation resided among populations, and only 6.00% could be attributed to variation within populations. A higher genetic differentiation of Q. infectoria populations was found according to cpSSR data in comparison to nSSR data. Cophenetic correlation coefficient values were statistically insignificant between nSSR and cpSSR data. The unweighted pair group method with arithmetic mean and Bayesian cluster analyses grouped the studied individuals into two main clusters based on both nSSR and cpSSR data. nSSR data could not completely clustered individuals next each other according to their geographical collection area. Information detailed by nSSR loci revealed that north-Zagros gall oak preserves average levels of genetic diversity at the species level, high level of within-population genetic diversity, and moderate level of genetic variation among populations. The present results provide valuable data for in situ or ex situ conservation and utilization of the studied germplasm.




Similar content being viewed by others
References
Aldrich PR, Michler CH, Sun WL, Romero-Severson J (2002) Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol Ecol Notes 2:472–474
Birky CW (1988) Evolution and variation in plant chloroplast and mitochondrial genomes. In: Gottlieb L, Jain S (eds) Plant Evolutionary Biology. Chapman and Hall, London, pp 23–53
Birky CW (1995) Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Nat Acad Sci 92(25):11331–11338
Bruschi P, Vendramin GG, Bussotti F, Grossoni P (2000) Morphological and molecular differentiation between Quercus petraea (Matt.) Liebl. and Quercus pubescens Willd. (Fagaceae) in Northern and Central Italy. Ann Bot 85:325–333
Deguilloux MF, Dumolin-Lapègue S, Gielly L, Grivet D, Petit RJ (2003) A set of primers for the amplification of chloroplast microsatellites in Quercus. Mol Ecol Notes 3:24–27
Dodd RS, Afzal-Rafii Z, Mayer W (2008) Molecular markers show how pollen and seed dispersal affect population genetic structure in Coast Live Oak (Quercus agrifolia Née). In: Standiford RB (ed) Proceedings of the sixth symposium on oak woodlands: today’s challenges, tomorrow’s opportunities. Pacific Southwest Research Station, Forest Service, US Department of Agriculture, Albany
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Ducousso A, Michaud H, Lumaret R (1993) Reproduction and gene flow in the genus Quercus L. Ann Sci For 50:91–106
Duminil J, Fineschi S, Hampe A, Jordano P, Salvini D, Vendramin GG, Petit RJ (2007) Can population genetic structure be predicted from life-history traits? Am Nat 169:662–672
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256
Echt CS, DeVerno L, Anzidei M, Vendramin GG (1998) Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Mol Ecol 7:307–316
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14(8):2611–2620
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7(4):574–578
FAO (2007) forestry in Iran, online, viewed 18 February 2008.
Grivet D, Sork VL, Westfall RD, Davis FW (2008) Conserving the evolutionary potential of California valley oak (Quercus lobata Née): a multivariate approach to conservation planning. Mol Ecol 17:139–156
Harris SA, Ingram R (1991) Chloroplast DNA and Biosystematics: The effects of intraspecific diversity and plastid transmission. Taxon 40(3):393
Henareh Khalyani A, Mayer AL (2013) Spatial and temporal deforestation dynamics of Zagros forests (Iran) from 1972 to 2009. Landsc Urban Plan 117:1–12
Heuertz M, Ois Hausman J-F, Hardy OJ, Vendramin GG, Frascaria-Lacoste N, Vekemans X (2004) Nuclear microsatellites reveal contrasting patterns of genetic structure between western and southeastern European populations of the common ash (Fraxinus excelsior L.) Evolution 58:976–988
Horvath A, Christmann H, Laigret F (2008) Genetic diversity and relationships among Prunus cerasifera (cherry plum) clones. Botany 86:1311–1318
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177(3):309–334
Kanowski P (1999) Forest and biological diversity. Paper presented at the training course on in situ conservation of forest genetic resources and rehabilitation of biodiversity, 23 August-September, Bangkok, Thailand
Khadivi-Khub A, Zamani Z, Fattahi R, Wünsch A (2014) Genetic variation in wild Prunus L. subgen. Cerasus germplasm from Iran characterized by nuclear and chloroplast SSR markers. Trees 28:471–485
Lahtinen MJ, Pulkkinen P, Helander ML (1996) Potential gene flow by pollen between English oak (Quercus robur L.) stands in Finland. For Stud 28:47–50
Lande R (1995) Mutation and Conservation. Conserv Biol 9(4):782–791
Lepais O, Petit R, Guichoux E, Lavabre J, Alberto F et al (2009) Species relative abundance and direction of introgression in oaks. Mol Ecol 18:2228–2242
Li J, Ge XJ, Cao HL, Ye WH (2007) Chloroplast DNA diversity in Castanopsis hystrix populations in south China. For Ecol Manag 243(1):94–101
Lind J, Gailing O (2013) Genetic structure of Quercus rubra L. and Quercus ellipsoidalis E. J. Hill populations at gene-based EST-SSR and nuclear SSR markers. Tree Genet Genomes 9:707–722
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
McCauley DE (1995) The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trend Ecol Evol 10(5):198–202
Mohanty A, Martin JP, Gonzaalez LM, Aguinagaldo I (2003) Association between chloroplast DNA and mitochondrial DNA haplotypes in Prunus spinosa L. (Rosaceae) populations across Europe. Ann Bot 92(6):749–755
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321–3323
Olfat OA, Pourtahmasi K (2010) Anatomical characters in three oak species (Q. libani, Q. brantii and Q. infectoria) from Iranian Zagros Mountains. Aus J Bas App Sci 4:3230–3237
Ouborg NJ, Piquot Y, Van Groenendael JM (1999) Population genetics, molecular markers and the study of dispersal in plants. J Ecol 87(4):551–568
Pakkad G, Ueno S, Yoshimaru H (2008) Genetic diversity and differentiation of Quercus semiserrata Roxb. In northern Thailand revealed by nuclear and chloroplast microsatellite markers. For Ecol Manag 255:1067–1077
Palmer JD (1987) Chloroplast DNA Evolution and Biosystematic Uses of Chloroplast DNA Variation. Amer Nat 130:S6–S29
Panahi P, Jamzad Z, Pourmajidian MR, Fallah A, Pourhashemi M, Sohrabi H (2012) Taxonomic revision of the Quercus brantii complex (Fagaceae) in Iran with emphasis on leaf and pollen micromorphology. Acta Bot Hung 54:355–375
Pavel AB, Vasile CI (2012) PyElph—a software tool for gel images analysis and phylogenetics. MC Bioinformat 13:9
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mole Ecol Note 6:288–295
Petit RJ, AEl M, Pons O (1998) Identifying Populations for Conservation on the Basis of Genetic Markers. Conserv Biol 12(4):844–855
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brever S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller Stack G, Demesure-Musch B, Palmé A, Martín JP, Rendell S, Vendramin G (2003) Glacial refugia: hotspots but no melting pots of genetic diversity. Science 300:1563–1565
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689–701
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147
Rahmani MS, Alikhani L, Shabanian N, Khadivi-Khub A (2015) Genetic differentiation in Quercus infectoria from northwest of Iran revealed by different nuclear markers. Tree Genet Genomes 11:800
Rajora OP (1999) Genetic biodiversity impacts of silvicultural practices and phenotypic selection in white spruce. Theor Appl Genet 99(6):954–961
Rajora OP, Mosseler A (2001) Challenges and opportunities for conservation of forest genetic resources. Euphytica 118:197–212
Robledo-Arnuncio JJ, Gil L (2005) Patterns of pollen dispersal in a small population of Pinus sylvestris L. revealed by total-exclusion paternity analysis. Heredity 94:13–22
Rohlf FJ (2000) NTSYS-pc numerical taxonomy and multivariate analysis system. Version 2.1. Exeter Software, Setauket
Sánchez-Ortiz K (2012) Estructura y diversidad genética de Quecus glabrescens a través de un gradiente de encinos blancos asociados. BsSC dissertation, Universidad Autónoma del Estado de Morelos
Schlötterer C, Pemberton J (1998) The use of microsatellites for genetic analysis of natural populations—a critical review. In: DeSalle R, Schierwater B (eds) Molecular approaches to ecology and evolution. Birkhaüser, Basel, pp 71–86
Schneider S, Kueffer J.M, Roessli D, Excoffier L (1997). ARLEQUIN. A software for population genetic data analysis, version 1.1. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Switzerland
Setsuko S, Ishida K, Ueno S, Tsumura Y, Tomaru N (2007) Population differentiation and gene flow within a metapopulation of a threatened tree, Magnolia stellata (Magnoliaceae). Am J Bot 94:128–136
Steinkellner H, Fluch S, Turetschek E, Lexer C, Streiff R, Kremer A, Burg K, Glossl J (1997) Identification and characterization of (GA/CT) n-microsatellite loci from Quercus petraea. Plant Mol Biol 33:1093–1096
Streiff R, Labbe T, Baculieri R, Steinkellner H, Glössl J, Kremer A (1998) Within-population genetic structure in Quercus robur L. and Quercus petraea (Matt.) Liebl. assessed with isozymes and microsatellites. Mol Ecol 7:317–328
Streiff R, Ducousso A, Lexer C, Steinkellner H, Gloessl J et al (1999) Pollen dispersal inferred from paternity analysis in a mixed stand of Quercus robur L. and Quercus petraea (Matt.) Liebl. Mol Ecol 8:831–841
Sullivan AR, Lind JF, McCleary TS, Romero-Severson J, Gailing O (2013) Development and characterization of genomic and gene-based microsatellite markers in North American red oak species. Plant Mol Biol Rep 31:231–239
Ueno S, Setsuko S, Kawahara T, Yoshimaru H (2006) Genetic diversity and differentiation of the endangered Japanese endemic tree Magnolia stellata using nuclear and chloroplast microsatellite markers. Conserv Genet 6(4):563–574
Valencia-Cuevas L, Piñero D, Mussali-Galante P, Valencia-Ávalos S, Tovar-Sánchez E (2014) Effect of a red oak species gradient on genetic structure and diversity of Quercus castanea (Fagaceae) in Mexico. Tree Genet Genomes 10:641–652
Weising K, Gardner RC (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42(1):9–19
Whitham TG, Bailey JK, Scheweitzer JA, Shuster SM, Bangert RK, LeRoy CJ, Lonsdorf EV, Allan GJ, DiFazio SP, Potts BM, Fischer DC, Gehrig CA, Lindroth RL, Marks JC, Hart SC, Wimp GM, Wooley SC (2006) A framework for community and ecosystem genetics: from genes to ecosystems. Nature 7:510–523
Wimp GM, Young PW, Woolbright SA, Martinsen GD, Keim P, Whitham TG (2004) Conserving plant genetic diversity for dependent animal communities. Ecol Lett 7:776–780
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A 84:9054–9058
Zhang R, Hipp AL, Gailing O (2015) Sharing of chloroplast haplotypes among red oak species suggests interspecific gene flow between neighboring populations. Botany, 2015 93(10):691–700
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Kremer
Electronic supplementary material
ESM 1
(XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Mohammad-Panah, N., Shabanian, N., Khadivi, A. et al. Genetic structure of gall oak (Quercus infectoria) characterized by nuclear and chloroplast SSR markers. Tree Genetics & Genomes 13, 70 (2017). https://doi.org/10.1007/s11295-017-1146-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1146-8