Abstract
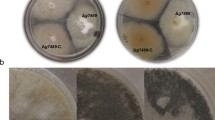
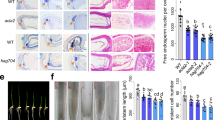
Core histones in the nucleosome are subject to a wide variety of posttranslational modifications (PTMs), such as methylation, phosphorylation, ubiquitylation, and acetylation, all of which are crucial in shaping the structure of the chromatin and the expression of the target genes. A putative histone methyltransferase LaeA/Lae1, which is conserved in numerous filamentous fungi, functions as a global regulator of fungal growth, virulence, secondary metabolite formation, and the production of extracellular glycoside hydrolases (GHs). LaeA’s direct histone targets, however, were not yet recognized. Previous research has shown that LaeA interacts with core histone H2B. Using S-adenosyl-l-methionine (SAM) as a methyl group donor and recombinant human histone H2B as the substrate, it was found that Penicillium oxalicum LaeA can transfer the methyl groups to the C-terminal lysine (K) 108 and K116 residues in vitro. The H2BK108 and H2BK116 sites on recombinant histone correspond to P. oxalicum H2BK122 and H2BK130, respectively. H2BK122A and H2BK130A, two mutants with histone H2B K122 or K130 mutation to alanine (A), were constructed in P. oxalicum. The mutants H2BK122A and H2BK130A demonstrated altered asexual development and decreased extracellular GH production, consistent with the findings of the laeA gene deletion strain (ΔlaeA). The transcriptome data showed that when compared to wild-type (WT) of P. oxalicum, 38 of the 47 differentially expressed (fold change ≥ 2, FDR ≤ 0.05) genes that encode extracellular GHs showed the same expression pattern in the three mutants ΔlaeA, H2BK122A, and H2BK130A. The four secondary metabolic gene clusters that considerably decreased expression in ΔlaeA also significantly decreased in H2BK122A or H2BK130A. The chromatin of promotor regions of the key cellulolytic genes cel7A/cbh1 and cel7B/eg1 compacted in the ΔlaeA, H2BK122A, and H2BK130A mutants, according to the results of chromatin accessibility real-time PCR (CHART-PCR). The chromatin accessibility index dropped. The histone binding pocket of the LaeA-methyltransf_23 domain is compatible with particular histone H2B peptides, providing appropriate electrostatic and steric compatibility to stabilize these peptides, according to molecular docking. The findings of the study demonstrate that H2BK122 and H2BK130, which are histone targets of P. oxalicum LaeA in vitro, are crucial for fungal conidiation, the expression of gene clusters encoding secondary metabolites, and the production of extracellular GHs.







Similar content being viewed by others
Data availability
The Whole Genome Shotgun projects have been deposited in DDBJ/EMBL/GenBank under the accession number AGIH00000000 (https://www.ncbi.nlm.nih.gov/bioproject/?term=AGIH00000000). The mass spectrometry has been deposited to the iProX (www.iprox.org) with ID: IPX0003226002. The raw data of expression profiling sequencing have been deposited in NCBI’s Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). The accession number of ΔlaeA, H2BK122A, and H2BK130A is GSE142715 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142715).
References
Amare MG, Keller NP (2014) Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet Biol 66:11–18
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7(10):986–995
Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320(5882):1504–1506
Beck HC, Nielsen EC, Matthiesen R, Jensen LH, Sehested M, Finn P, Grauslund M, Hansen AM, Jensen ON (2006) Quantitative proteomic analysis of post-translational modifications of human histones. Mol Cell Proteomics 5(7):1314–1325
Binda O (2013) On your histone mark. SET Epigenet 8(5):457–463
Black JC, Van Rechem C, Whetstine JR (2012) Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48(4):491–507
Bok JW, Keller NP (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3(2):527–535
Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Keller NP (2005) LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell 4(9):1574–1582
Cea-Sanchez S, Corrochano-Luque M, Gutierrez G, Glass NL, Canovas D, Corrochano LM (2022) Transcriptional regulation by the ve0lvet protein VE-1 during asexual development in the fungus Neurospora crassa. Mbio. https://doi.org/10.1128/mbio.01505-22
Chandrasekharan MB, Huang F, Chen YC, Sun ZW (2010) Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol Cell Biol 30(13):3216–3232
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676
Connolly LR, Smith KM, Freitag M (2013) The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet 9(10):e1003916
Croken MM, Nardelli SC, Kim K (2012) Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol 28(5):202–213
Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW (2011) Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7(2):113–119
Fuchs G, Oren M (2014) Writing and reading H2B monoubiquitylation. Biochim Biophys Acta 1839(8):694–701
Fulton MD, Brown T, Zheng YG (2018) Mechanisms and inhibitors of histone arginine methylation. Chem Rec 18(12):1792–1807
Gacek-Matthews A, Berger H, Sasaki T, Wittstein K, Gruber C, Lewis ZA, Strauss J (2016) KdmB, a jumonji histone H3 demethylase, regulates genome-wide H3K4 trimethylation and is required for normal induction of secondary metabolism in Aspergillus nidulans. PLoS Genet 12(8):e1006222
Gauthier GM, Keller NP (2013) Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol 61:146–157
Hao L, Zhang M, Yang C, Pan X, Wu D, Lin H, Ma D, Yao Y, Fu W, Chang J, Yang Y, Zhuang Z (2023) The epigenetic regulator Set9 harmonizes fungal development, secondary metabolism, and colonization capacity of Aspergillus flavus. Int J Food Microbiol 403:110298
Hayashi-Takanaka Y, Kina Y, Nakamura F, Becking LE, Nakao Y, Nagase T, Nozaki N, Kimura H (2020) Histone modification dynamics as revealed by multicolor immunofluorescence-based single-cell analysis. J Cell Sci. https://doi.org/10.1242/jcs.243444
Janevska S, Baumann L, Sieber CMK, Munsterkotter M, Ulrich J, Kamper J, Guldener U, Tudzynski B (2018a) Elucidation of the two H3K36me3 histone methyltransferases Set2 and Ash1 in Fusarium fujikuroi unravels their different chromosomal targets and a major impact of ash1 on genome stability. Genetics 208(1):153–171
Janevska S, Guldener U, Sulyok M, Tudzynski B, Studt L (2018b) Set1 and Kdm5 are antagonists for H3K4 methylation and regulators of the major conidiation-specific transcription factor gene ABA1 in Fusarium fujikuroi. Environ Microbiol 20(9):3343–3362
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293(5532):1074–1080
Judes G, Dagdemir A, Karsli-Ceppioglu S, Lebert A, Echegut M, Ngollo M, Bignon YJ, Penault-Llorca F, Bernard-Gallon D (2016) H3K4 acetylation, H3K9 acetylation and H3K27 methylation in breast tumor molecular subtypes. Epigenomics 8(7):909–924
Kim HK, Lee S, Jo SM, McCormick SP, Butchko RA, Proctor RH, Yun SH (2013a) Functional roles of FgLaeA in controlling secondary metabolism, sexual development, and virulence in Fusarium graminearum. PLoS ONE 8(7):e68441
Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD, Roeder RG (2013b) The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell 49(6):1121–1133
Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25(8):3305–3316
Krappmann S, Bayram O, Braus GH (2005) Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryot Cell 4(7):1298–1307
Kuhad RC, Gupta R, Singh A (2011) Microbial cellulases and their industrial applications. Enzyme Res 2011:280696
Lee KY, Ranger M, Meneghini MD (2018) Combinatorial genetic control of Rpd3S through histone H3K4 and H3K36 methylation in budding yeast. G3 (bethesda). https://doi.org/10.1534/g3.118.200589
Li B, Howe L, Anderson S, Yates JR 3rd, Workman JL (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278(11):8897–8903
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967
Li Y, Zheng X, Zhang X, Bao L, Zhu Y, Qu Y, Zhao J, Qin Y (2016) The different roles of Penicillium oxalicum LaeA in the production of extracellular cellulase and beta-xylosidase. Front Microbiol 7:2091
Li Y, Hu Y, Zhu Z, Zhao K, Liu G, Wang L, Qu Y, Zhao J, Qin Y (2019) Normal transcription of cellulolytic enzyme genes relies on the balance between the methylation of H3K36 and H3K4 in Penicillium oxalicum. Biotechnol Biofuels 12:198
Li Y, Zhao L, Zhang Y, Wu P, Xu Y, Mencius J, Zheng Y, Wang X, Xu W, Huang N, Ye X, Lei M, Shi P, Tian C, Peng C, Li G, Liu Z, Quan S, Chen Y (2022) Structural basis for product specificities of MLL family methyltransferases. Mol Cell. https://doi.org/10.1016/j.molcel.2022.08.022
Liu G, Zhang L, Wei X, Zou G, Qin Y, Ma L, Li J, Zheng H, Wang S, Wang C, Xun L, Zhao GP, Zhou Z, Qu Y (2013) Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS ONE 8(2):e55185
Lu S, Xie YM, Li X, Luo J, Shi XQ, Hong X, Pan YH, Ma X (2009) Mass spectrometry analysis of dynamic post-translational modifications of TH2B during spermatogenesis. Mol Hum Reprod 15(6):373–378
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 389(6648):251–260
Luo Q, Li N, Xu JW (2022) A methyltransferase LaeA regulates ganoderic acid biosynthesis in Ganoderma lingzhi. Front Microbiol 13:1025983
Martin JF (2017) Key role of LaeA and velvet complex proteins on expression of beta-lactam and PR-toxin genes in Penicillium chrysogenum: cross-talk regulation of secondary metabolite pathways. J Ind Microbiol Biotechnol 44(4–5):525–535
Martin BJE, Brind’Amour J, Kuzmin A, Jensen KN, Liu ZC, Lorincz M, Howe LJ (2021) Transcription shapes genome-wide histone acetylation patterns. Nat Commun 12(1):210
Mayorga ME, Timberlake WE (1990) Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics 126(1):73–79
Mello-de-Sousa TM, Gorsche R, Rassinger A, Pocas-Fonseca MJ, Mach RL, Mach-Aigner AR (2014) A truncated form of the carbon catabolite repressor 1 increases cellulase production in Trichoderma reesei. Biotechnol Biofuels 7(1):129
Mello-de-Sousa TM, Rassinger A, Pucher ME, dos Santos Castro L, Persinoti GF, Silva-Rocha R, Pocas-Fonseca MJ, Mach RL, Nascimento Silva R, Mach-Aigner AR (2015) The impact of chromatin remodelling on cellulase expression in Trichoderma reesei. BMC Genomics 16:588
Millan-Zambrano G, Burton A, Bannister AJ, Schneider R (2022) Histone post-translational modifications—cause and consequence of genome function. Nat Rev Genet 23(9):563–580
Palmer JM, Keller NP (2010) Secondary metabolism in fungi: does chromosomal location matter? Curr Opin Microbiol 13(4):431–436
Patananan AN, Palmer JM, Garvey GS, Keller NP, Clarke SG (2013) A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine. J Biol Chem 288(20):14032–14045
Pinto D, Page V, Fisher RP, Tanny JC (2021) New connections between ubiquitylation and methylation in the co-transcriptional histone modification network. Curr Genet 67(5):695–705
Qin Y, Ortiz-Urquiza A, Keyhani NO (2014) A putative methyltransferase, mtrA, contributes to development, spore viability, protein secretion and virulence in the entomopathogenic fungus Beauveria bassiana. Microbiology 160(Pt 11):2526–2537
Raduwan H, Isola AL, Belden WJ (2013) Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 protein is needed for normal clock gene expression. J Biol Chem 288(12):8380–8390
Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J (2010) Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol 76(6):1376–1386
Robzyk K, Recht J, Osley MA (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287(5452):501–504
Sankar A, Mohammad F, Sundaramurthy AK, Wang H, Lerdrup M, Tatar T, Helin K (2022) Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat Genet 54(6):754–760
Sarikaya Bayram O, Bayram O, Valerius O, Park HS, Irniger S, Gerke J, Ni M, Han KH, Yu JH, Braus GH (2010) LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet 6(12):e1001226
Sarikaya-Bayram O, Palmer JM, Keller N, Braus GH, Bayram O (2015) One Juliet and four Romeos: VeA and its methyltransferases. Front Microbiol 6:1
Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, Kubicek CP (2012) The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol 84(6):1150–1164
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403(6765):41–45
Strauss J, Reyes-Dominguez Y (2011) Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet Biol 48(1):62–69
Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18(1):207–208
Sugui JA, Pardo J, Chang YC, Mullbacher A, Zarember KA, Galvez EM, Brinster L, Zerfas P, Gallin JI, Simon MM, Kwon-Chung KJ (2007) Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot Cell 6(9):1552–1561
Sun X, Liu Z, Qu Y, Li X (2008) The effects of wheat bran composition on the production of biomass-hydrolyzing enzymes by Penicillium decumbens. Appl Biochem Biotechnol 146(1–3):119–128
Uckelmann M, Sixma TK (2017) Histone ubiquitination in the DNA damage response. DNA Repair (amst) 56:92–101
Venkatesh S, Workman JL (2013) Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development. Wiley Interdiscip Rev Dev Biol 2(5):685–700
Vogel. (1956) A convenient growth medium for Neurospora (Medium N). Microbial Genet Bull 13:42–43
Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13(2):115–126
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362
Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28(4):1348–1360
Yin S, Yang D, Zhu Y, Huang B (2022) Methionine and S-adenosylmethionine regulate monascus pigments biosynthesis in Monascus purpureus. Front Microbiol 13:921540
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41(11):973–981
Zhang X, Qu Y, Qin Y (2016a) Expression and chromatin structures of cellulolytic enzyme gene regulated by heterochromatin protein 1. Biotechnol Biofuels 9(1):206
Zhang X, Zhu Y, Bao L, Gao L, Yao G, Li Y, Yang Z, Li Z, Zhong Y, Li F, Yin H, Qu Y, Qin Y (2016b) Putative methyltransferase LaeA and transcription factor CreA are necessary for proper asexual development and controlling secondary metabolic gene cluster expression. Fungal Genet Biol 94:32–46
Zhang C, Zhang H, Zhu Q, Hao S, Chai S, Li Y, Jiao Z, Shi J, Sun B, Wang C (2020a) Overexpression of global regulator LaeA increases secondary metabolite production in Monascus purpureus. Appl Microbiol Biotechnol 104(7):3049–3060
Zhang X, Li M, Zhu Y, Yang L, Li Y, Qu J, Wang L, Zhao J, Qu Y, Qin Y (2020b) Penicillium oxalicum putative methyltransferase Mtr23B has similarities and differences with LaeA in regulating conidium development and glycoside hydrolase gene expression. Fungal Genet Biol 143:103445
Zhang X, Hu Y, Liu G, Liu M, Li Z, Zhao J, Song X, Zhong Y, Qu Y, Wang L, Qin Y (2022) The complex Tup1-Cyc8 bridges transcription factor ClrB and putative histone methyltransferase LaeA to activate the expression of cellulolytic genes. Mol Microbiol. https://doi.org/10.1111/mmi.14885
Zhao Z, Gu S, Liu D, Liu D, Chen B, Li J, Tian C (2023) The putative methyltransferase LaeA regulates mycelium growth and cellulase production in Myceliophthora thermophila. Biotechnol Biofuels Bioprod 16(1):58
Acknowledgements
We thank Xiuyun Wu of the State Key Laboratory of Microbial Technology in Shandong University for her help in nucleosome structure analyses.
Funding
This research was funded by National Natural Sciences Foundation of China (32070077 and 32370075), the Shandong Provincial Natural Science Foundation (ZR2020QC003), and China Postdoctoral Science Foundation (2020M682170).
Author information
Authors and Affiliations
Contributions
XZ wrote the main manuscript text, YY prepared seven figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Yang, Y., Wang, L. et al. Histone H2B lysine 122 and lysine 130, as the putative targets of Penicillium oxalicum LaeA, play important roles in asexual development, expression of secondary metabolite gene clusters, and extracellular glycoside hydrolase synthesis. World J Microbiol Biotechnol 40, 179 (2024). https://doi.org/10.1007/s11274-024-03978-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-024-03978-0