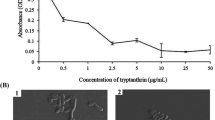
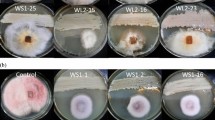
Abstract
A strain of Bacillus amyloliquefaciens (VCRC B483) exhibiting mosquito pupicidal, keratinase and antimicrobial activities was isolated from mangrove forest ecosystem of Andaman and Nicobar Islands. Molecular characterization of the strain showed the presence of lipopeptide encoding bmyC gene. Phylogenetic tree based on protein sequence of this gene exhibited homology with mycosubtilin synthetase of Bacilus atropheus and Iturin synthetase of Bacillus subtilis and B. amyloliquefaciens. This is the first report on the evolutionary conservation of amino acids concerned with the function and structure of bmyC protein of B. amyloliquefaciens. The presence of valine at the 1197th position in our strain was found to be unique and different from the existing strains of B. subtilis and B. amyloliquefaciens. Molecular modelling studies revealed significant changes in the structure of epimerization domain of the bmyC protein with A1197V variation. Crude metabolite of this strain exhibited antifungal activity against Fusarium sp. and Carvularia sp.








Similar content being viewed by others
References
Alvarez F, Castro M, Principe A, Borioli G, Fischer S, Mori G, Jofre E (2011) The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J Appl Microbiol 112:159–174
Ashkenzay H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) Consurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38:W529W533
Athukorala SNP, Dilantha Fernando WG, Rashid KY (2009) Identification of antifungal antibiotics of Bacillus species isolated from different microhabitats using polymerase chain reaction and MALDI-TOF mass spectrometry. Can J Microbiol 55:1021–1032
Basha S, Ulaganathan K (2002) Antagonism of Bacillus sp. BC121 towards Curvularia lunata. Curr Sci 82:1457–1463
Benitez LB, Velho RV, Lisboa MP, Medina LF, Brandelli A (2010) Isolation and characterization of antifungal peptides produced by B amyloliquefaciens LBM5006. J Microbiol 48:791–797
Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N (2004) ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20:1322–1324
Caldeira AT, Santos Arteiro JM, Coelho AV, Roseiro JC (2011) Combined use of LC-ESI-MS and antifungal tests for rapid identification of bioactive lipopeptides produced by Bacillus amyloliquefaciens CCMI 1051. Process Biochem 46:1738–1746
Caradec T, Pupin M, Vanvlassenbroeck A, Devignes MD, Smail-Tabbone M, Jacques P, Leclere V (2014) Prediction of monomer isomery in florine: a workflow dedicated to Nonribosomal peptide discovery. PLoS ONE 9:e85667
Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N (2013) Consurf: using evolutionary data to raise testable hypotheses about protein function. Isr J Chem 53:199–206
Finking R, Marahiel MA (2004) Biosynthesis of nonribosomal peptides. Annu Rev Microbiol 58:453–488
Geetha I, Manonmani AM (2008) Mosquito pupicidal toxin production by Bacillus subtilis subsp. subtilis. Biol Cont 44:242–247
Geetha I, Manonmani AM, Prabakaran G (2011) Bacillus amyloliquefaciens: a mosquitocidal bacterium from mangrove forests of Andaman & Nicobar islands, India. Acta Trop 120:155–159
Gupta U, Banerjee K, Gabrani R, Gupta S, Sharma SK, Jain CK (2011) Variability analyses of functional domains within glucosamine-6-phosphate synthase of mycoses-causing fungi. Bioinformation 6:196–199
Han Y, Zhang B, Shen Q, You C, Yu Y, Li P, Shang Q (2015) Purification and identification of two antifungal cyclic peptides produced by Bacillus amyloliquefaciens L-H15. Appl Biochem Biotechnol 176:2202–2212
Ishihara H, Takoh M, Nishibayashi R, Sato A (2002) Distribution and variation of Bacitracin synthetase gene sequences in laboratory stock strains of Bacillus licheniformis. Curr Microbiol 45:18–23
Joshi R, Bharucha CH, Desai (2007) Production of biosurfactant and antifungal compound by fermented food isolate Bacillus subtilis 20B. Biores Technol 99:4603–4608
Koumautsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing non ribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186:1084–1096
Li B, Li Q, Xu Z, Zhang N, Shen Q, Zhang R (2014) Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front Microbiol 5:1–10
Mandlik V, Shinde S, Singh S (2014) Molecular evolution of the enzymes involved in the sphingolipid metabolism of Leishmania: selection pressure in relation to functional divergence and conservation. BMC Evol Biol 14:142
Moyne AL, Cleveland TE, Tuzun S (2004) Molecular characterization and analysis of operon encoding the antifungal lipopeptide bacillomycin D. FEMS Microbiol Lett 234:43–49
Ongena M, Jacques P (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16:116–125
Patel S, Mackerell AD, Brooks CL (2004) CHARMM fluctuating charge force field for proteins: II protein/solvent properties from molecular dynamics simulations using a nonadditive electrostatic model. J Comput Chem 25:1504–1514
Peypoux F, Besson F, Michel G (1980) Characterization of a new antiobiotic of iturin group: bacillomycin D. J Antibiot 10:1146–1149
Peypoux F, Besson F, Michel G, Delcambe L (1981) Structure of bacillomycin D, a new antibiotic of the iturin group. Eur J Biochem 118:323–327
Peypoux F, Pommier MT, Das BC, Besson F, Delcambe L, Michel G (1984) Structures of bacillomycin D and bacillomycin L peptidolipid antibiotics from Bacillus subtilis. J Antibiot 12:1600–1604
Ramarathnam R, Bo S, Chen Y, Dilantho Fernando WG, Xuewen G, De Kievit T (2007) Molecular and biochemical detection of fengycin and Bacillomycin D producing Bacillus spp., antagonistic to fungal pathogens of Canola and Wheat. Can J Microbiol 53:901–911
Shrivastava A, Gupta MK, Singhal PK (2013) Nutritional and environmental optimization of antifungal potential of Bacillus strains. IJAR 1:1–6
Stachelhaus T, Schneide RA, Marahiel MA (1995) Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science 269:69–72
Streiker M, Tanovic A, Marahiel MA (2010) Nonribosomal peptide synthetases: structure and dynamics. Curr Opin Struct Biol 20:234–240
Sumi CD, Yang BW, Yeo I, Hahm YT (2014) Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can J Microbiol 61:93–103
Tabbene O, Kalai L, Slimene IB, Karkouch I, Elkahoui S, Gharbi A, Cosette P, Mangoni M, Jouenne T, Limam F (2011) Anti-candida effect of bacillomycin D-like lipopeptides from Bacillus subtilis B38. FEMS Microbiol Lett 316:108–114
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tanaka K, Amaki Y, Ishihara A, Nakajima A (2015) Synergistic effects of surfactin homologues with bacillomycin D in suppression of gray mold disease by B amyloliquefaciens biocontrol strain SD-32. J Agri Food Chem 63:5344–5353
Xu Z, Shao J, Li B, Yan X, Shen Q, Zhang R (2013) Contribution of bacillomycin D in B amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl Environ Microbiol 79:808–815
Yu GY, Sinclair JB, Hartman GL, Bertagnolli BL (2002) Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol Biochem 34:955–963
Zhao P, Quan C, Jin L, Wang L, Guo X, Fan S (2013) Sequence characterization and computational analysis of the nonribosomal peptide synthetases controlling biosynthesis of lipopeptides, fengycin and bacillomycin D, from Bacillus amyloliquefaciens Q426. Biotechnol Lett 35:2155–2163
Acknowledgements
The authors are grateful to the Director, Vector Control Research Centre, Puducherry for his encouragement and valuable suggestions. Technical assistance rendered by Mr. S. Bhoopal Chakravarthy, Mrs. K. Vijayalakshmi and contribution of Mr. Y. Srinivas Murty in electronic artwork is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11274_2018_2498_MOESM1_ESM.pdf
Consurf color coded mutiple sequence alignment of partial bmyC protein (AHW47908) with 50 homologue sequences showing amino acid conservation (PDF 362 KB)
Rights and permissions
About this article
Cite this article
Ashokkumar, M., Irudayaraj, G., Yellapu, N. et al. Molecular characterization of bmyC gene of the mosquito pupicidal bacteria, Bacillus amyloliquefaciens (VCRC B483) and in silico analysis of bacillomycin D synthetase C protein. World J Microbiol Biotechnol 34, 116 (2018). https://doi.org/10.1007/s11274-018-2498-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-018-2498-4