Abstract
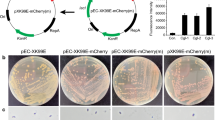
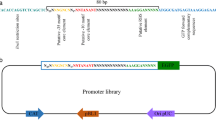
Available molecular and genetic tools for the genetic manipulation of Arthrobacter species are limited until now. In gene engineering, a continuous set of promoters with various strengths are of importance for fine-tuning gene expression in metabolic optimization and control analysis. Here, for the first time, we constructed a promoter trap system using green fluorescence protein (GFP) as a reporter, for screening and characterizing functional Arthrobacter promoters. Twenty-three Arthrobacter transformants of various GFP fluorescence strengths were isolated and characterized through the analysis of DNA sequences. Among the 23 putative promoters, 2 were selected for deletion analysis of promoter elements. As a result, the deletion of the upstream of the putative promoter P8 and P13 caused a 43.8% decrease and a 29.1% increase in the fluorescence signals, respectively. Finally, we obtained the strongest promoter P13-3 which was 4.4 times more potent than the promoter of 6–hydroxyl–d–nicotine oxidase gene which was previously reported in Arthrobacter nicotinovorans, and the obtained promoter was used to improve the production of cyclic adenosine monophosphate in Arthrobacter sp. CGMCC 3584. The screening strategy together with obtained promoters in this study would contribute to the future engineering of Arthrobacter species.






Similar content being viewed by others
References
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bugg TD, Ahmad M, Hardiman EM, Singh R (2011) The emerging role for bacteria in lignin degradation and bio-product formation Current. Opin Biotechnol 22:394
Chen M, Xiao X, Wang P, Zeng X, Wang F (2005) Arthrobacter ardleyensis sp. nov., isolated from Antarctic lake sediment and deep-sea sediment. Arch Microbiol 183:301–305
Chiribau CB, Sandu C, Igloi GL, Brandsch R (2005) Characterization of PmfR, the transcriptional activator of the pAO1-borne purU-mabO-folD operon of Arthrobacter nicotinovorans. J Bacteriol 187:3062–3070
Estrem ST, Ross W, Gaal T, Chen ZS, Niu W, Ebright RH, Gourse RL (1999) Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev 13:2134–2147
Ganas P, Igloi GL, Brandsch R (2009) The megaplasmid pAO1 of Arthrobacter nicotinovorans and nicotine catabolism. In: Schwartz E (eds) Microbial megaplasmids, vol 11. Springer, Berlin, pp 271–282
Gourse RL, Ross W, Gaal T (2000) UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol 37:687–695
Guddat LW, Vos S, Martin JL, Keough DT, De JJ (2002) Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase. Protein Sci 11:1626–1638
He Y et al (2012) Cloning, expression, and characterization of an adenylate cyclase from Arthrobacter sp. CGMCC 3584. Appl Microbiol Biotechnol 96:963–970
Iwai A et al (1994) Molecular cloning and expression of an isomalto-dextranase gene from Arthrobacter globiformis T6. J Bacteriol 176:7730–7734
Lu L (2004) Autoinducer-2-based quorum sensing response of Escherichia coli to subtherapeutic tetracycline exposure. http://oaktrust.library.tamu.edu/bitstream/handle/1969.1/4198/etd-tamu-2004B-FSTC-Lu.pdf
Malphettes L, Weber CC, El-Baba MD, Schoenmakers RG, Aubel D, Weber W, Fussenegger M (2005) A novel mammalian expression system derived from components coordinating nicotine degradation in Arthrobacter nicotinovorans pAO1. Nucleic Acids Res 33:e107
Mauch L, Bichler V, Brandsch R (1990) Functional analysis of the 5′ regulatory region and the UUG translation initiation codon of the Arthrobacter oxidans 6-hydroxy-d-nicotine oxidase gene. Mol Gen Genet MGG 221:427–434
Munch A, Stingl L, Jung K, Heermann AR (2008) Photorhabdus luminescens genes induced upon insect infection. BMC Genomics 9:229
Niewerth H, Parschat K, Rauschenberg M, Ravoo BJ, Fetzner S (2013) The PaaX-type repressor MeqR2 of Arthrobacter sp. strain Rue61a, involved in the regulation of quinaldine catabolism, binds to its own promoter and to catabolic promoters and specifically responds to anthraniloyl coenzyme A. J Bacteriol 195:1068–1080
Nishiya Y, Imanaka T (1996) Analysis of a negative regulator, SoxR, for the Arthrobacter sarcosine oxidase gene. J Ferment Bioeng 81:64–67
Niu H et al (2013) Metabolic flux analysis of Arthrobacter sp. CGMCC 3584 for cAMP production based on 13C tracer experiments and gas chromatography–mass spectrometry. J Biotechnol 168:355–361
Parschat K, Overhage J, Strittmatter AW, Henne A, Gottschalk G, Fetzner S (2007) Complete nucleotide sequence of the 113-kilobase linear catabolic plasmid pAL1 of Arthrobacter nitroguajacolicus Rü61a and transcriptional analysis of genes involved in quinaldine degradation. J Bacteriol 189:3855–3867
Rediers H, Rainey PB, Vanderleyden J, De Mot R (2005) Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol Mol Biol Rev 69:217–261
Ross W, Aiyar SE, Salomon J, Gourse RL (1998) Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol 180:5375–5383
Ruiz-Cruz S, Solano-Collado V, Espinosa M, Bravo A (2010) Novel plasmid-based genetic tools for the study of promoters and terminators in Streptococcus pneumoniae and Enterococcus faecalis. J Microbiol Methods 83:156–163
Sandu C, Chiribau CB, Sachelaru P, Brandsch R (2005) Plasmids for nicotine-dependent and-independent gene expression in Arthrobacter nicotinovorans and other Arthrobacter species. Appl Environ Microbiol 71:8920–8924
Seo JS, Keum YS, Li QX (2009) Bacterial degradation of aromatic compounds. Int J Environ Res Public Health 6:278–309
Yao Y, Tang H, Su F, Xu P (2015) Comparative genome analysis reveals the molecular basis of nicotine degradation and survival capacities of. Arthrobacter. Sci Rep 5:8642
Zaide G, Grosfeld H, Ehrlich S, Zvi A, Cohen O, Shafferman A (2011) Identification and characterization of novel and potent transcription promoters of Francisella tularensis. Appl Environ Microbiol 77:1608–1618
Zhang X, Fuller JH, Mcintire WS (1993) Cloning, sequencing, expression, and regulation of the structural gene for the copper/topa quinone-containing methylamine oxidase from Arthrobacter strain P1, a gram-positive facultative methylotroph. J Bacteriol 175:5617
Acknowledgements
This work was supported by the National High-Tech Research and Development Program of China (863) (2012AA021203), the National Basic Research Program of China (973) (2013CB733602), the Major Research Plan of the National Natural Science Foundation of China (21390204), the National Technology Support Program (2012BAI44G01), the National Natural Science Foundation of China, General Program (2137611), the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R28), the young investigator grant program of National Natural Science Foundation of China (21506097 and 21606128) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. All authors read and approved the final manuscript.
Rights and permissions
About this article
Cite this article
Niu, H., Yang, W., Zhuang, K. et al. Screening of promoters from Arthrobacter sp. CGMCC 3584 using a green fluorescent protein reporter system. World J Microbiol Biotechnol 33, 208 (2017). https://doi.org/10.1007/s11274-017-2375-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-017-2375-6