Abstract
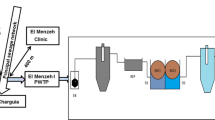
Biofilm formation on reverse osmosis (RO) systems represents a drawback in the application of this technology by different industries, including oil refineries. In RO systems the feed water maybe a source of microbial contamination and thus contributes for the formation of biofilm and consequent biofouling. In this study the planktonic culturable bacterial community was characterized from a feed water of a RO system and their capacities were evaluated to form biofilm in vitro. Bacterial motility and biofilm control were also analysed using phages. As results, diverse Protobacteria, Actinobacteria and Bacteroidetes were identified. Alphaproteobacteria was the predominant group and Brevundimonas, Pseudomonas and Mycobacterium the most abundant genera. Among the 30 isolates, 11 showed at least one type of motility and 11 were classified as good biofilm formers. Additionally, the influence of non-specific bacteriophage in the bacterial biofilms formed in vitro was investigated by action of phages enzymes or phage infection. The vB_AspP-UFV1 (Podoviridae) interfered in biofilm formation of most tested bacteria and may represent a good alternative in biofilm control. These findings provide important information about the bacterial community from the feed water of a RO system that may be used for the development of strategies for biofilm prevention and control in such systems.






Similar content being viewed by others
References
Abee T, Kovács AT, Kuipers OP, van der Veen S (2011) Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179
Ahiwale S, Tamboli N, Thorat K, Kulkarni R, Ackermann H, Kapadnis B (2011) In vitro management of hospital Pseudomonas aeruginosa biofilm using indigenous T7-like lytic phage. Curr Microbiol 62:335–340
Ainsworth S, Zomer A, Mahony J, van Sinderen D (2013) Lytic infection of Lactococcus lactis by bacteriophages Tuc 2009 and c2 triggers alternative transcriptional host responses. Appl Environ Microbiol 79(16):4786–4798
Allison DG (2003) The biofilm matrix. Biofouling 19:139–150
Azeredo J, Sutherland IW (2008) The use of phages for the removal of infectious biofilms. Curr Pharm Biotechnol 9:261–266
Bereschenko LA, Stams AJM, Heilig GHJ, Euverink GJW, Nederlof MM, Van Loosdrecht MCM (2007) Investigation of microbial communities on reverse osmosis membranes used for process water production. Water Sci Technol 55:181–190
Bereschenko LA, Heilig GHJ, Nederlof MM, van Loosdrecht MCM, Stams AJM, Euverink GJW (2008) Molecular characterization of the bacterial communities in the different compartments of a full-scale reverse-osmosis water purification plant. Appl Environ Microbiol 74:5297–5304
Bereschenko LA, Stams AJM, Wuverink GJW, van Loosdrecht MCM (2010) Biofilm formation on reverse osmosis membranes is initiated and dominated by Sphingomonas spp. Appl Environ Microbiol 76:2623–2632
Boles BR, Thoendel M, Singh PK (2005) Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol 57:1210–1223
Burdman S, Okon Y, Jurkevitch E (2000) Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Crit Rev Microbiol 26:91–110
Caiazza NC, Merrit JH, Brothers KM, O’Toole GA (2007) Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612
Chen CL, Liu WT, Chong ML, Wong MT, Ong SL, Seah H, Ng WJ (2004) Community structure of microbial biofilms associated with membrane-based water purification processes as revealed using a polyphasic approach. Appl Microbiol Biotechnol 63:466–473
Chen Y, Golding I, Sawai S, Guo L, Cox EC (2005) Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol 3(7):e229
Clark JR, March JB (2006) Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol 24(5):212–218
Cornelissen A, Ceyssens PJ, T’Syen J, Van Praet H, Noben JP, Shaburova OV, Krylov VN, Volckaert G, Lavigne R (2011) The T7-related Pseudomonas putida phage phi15 displays virion-associated biofilm degradation properties. PLoS One 6(4):e18597
Déziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by 57RP correlates Pseudomonas aeruginosa correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183:1195
Dias RS, Eller MR, Duarte VS, Pereira ÂL, Silva CC, Mantovani HC, Oliveira LL, Silva AM, De Paula SO (2013) Use of phages against antibiotic-resistant Staphylococcus aureus isolated from bovine mastitis. J Anim Sci 91:3930–3939
Eguchi M, Ostrowski M, Fegatella F, Bowman J, Nichols D, Nishino T, Cavicchioli R (2001) Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl Environ Microbiol 67:4945–4954
Eller MR, Dias RS, Moraes CA, Carvalho AF, Oliveira LL, Silva EA, Silva CC, De Paula SO (2012) Molecular characterization of a new lytic bacteriophage isolated from cheese whey. Arch Virol 157:2265–2272
Ewing B, Hillier L, Wendl MC, Green P (1998) Base calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Extremina CI, Costa L, Aguiar AI, Peixe L, Fonseca AP (2011) Optimization of processing conditions for the quantification of enterococci biofilms using microtitre-plates. J Microbiol Methods 84:167–173
Fu W, Forster T, Mayer O, Curtin JJ, Lehman SM, Donlan RM (2010) Bacteriophage cocktail for the prevention of biofilm formation by Pseudomonas aeruginosa on catheters in an in vitro model system. Antimicrob Agents Chemother 54:397–404
Goldman G, Starosvetsky J, Armon R (2009) Inhibition of biofilm on UF membrane by use of specific bacteriophages. J Membr Sci 342:145–152
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Gutierrez D, Martín-Platero AM, Rodriguez A, Martínez-Bueno M, García P, Martínez B (2011) Typing of bacteriophages by randomly amplified polymorphic DNA (RAPD)-PCR to assess genetic diversity. FEMS Microbiol Lett 322:90–97
Houry A, Briandet R, Aymerich S, Gohar M (2010) Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156:1009–1018
Huang LN, De Wever H, Diels L (2008) Diverse and distinct bacterial communities induced biofilm fouling in membrane bioreactors operated under different conditions. Environ Sci Technol 42:8360–8366
Hughes KA, Sutherland IW, Clark J, Jones MV (1998a) Bacteriophage and associated polysaccharides depolymerases—novel tools for study of bacterial biofilms. J Appl Microbiol 85:583–590
Hughes KA, Sutherland IW, Jones MV (1998b) Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology 144:3039–3047
Ivnitsky H, Katz I, Minz D, Volvovic G, Shimoni E, Kesselman E, Semiat R, Dosoretz CG (2007) Bacterial community composition and structure of biofilms developing on nanofiltration membranes applied to wastewater treatment. Water Res 41:3924–3935
Kelly D, McAuliffe O, Ross RP, Coffey A (2012) Prevention of Staphylococcus aureus biofilm formation and reduction in established biofilm density using a combination of phage K and modified derivates. Lett Appl Microbiol 54:286–291
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirov SM, Castrisios M, Shaw JG (2004) Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect Immun 72:1939–1945
Lemon KP, Higgins DE, Kolter R (2007) Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol 189:4418–4424
Melo LF, Flemming HC (1993) Mechanistic aspects of heat exchanger and membrane biofouling and prevention. In: Amjad Z (ed) Reverse osmosis: membrane technology, water chemistry and industrial applications. CDC Press, New York, p 365–380
Momba MNB, Kfir R, Venter SN, Loete TEC (2000) An overview of biofilm formation in distribution systems and its impact on the deterioration of water quality. Water SA 26:59–66
Murray TS, Kazmierczak BI (2008) Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708
Nguyen T, Roddick FA, Fan L (2012) Biofouling of water treatment membranes: a review of the underlying causes, monitoring techniques and control measures. Membranes 2:804–840
O’Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp 30:47
O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461
O’Toole GA, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156
Pollock TJ, Armentrout RW (1999) Planktonic/sessile dimorphism of polysaccharide-encapsulated sphingomonads. J Ind Microbiol Biotechnol 23:436–441
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Ridgway HF, Safarik J (1991) Biofouling of reverse osmosis membranes. In: Flemming H-C, Geesey GG (eds) Biofouling and biocorrosion in industrial water system. Springer, Heidelberg, pp 81–111
Rohban R, Amoozegar MA, Ventosa A (2009) Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake Iran. J Ind Microbiol Biotechnol 36:333–340
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salahi A, Mohammadi T, Rekabdar F, Mahdavi H (2010) Reverse osmosis of refinery oily wastewater effluents. Iran J Environ Health 7:413–422
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Schafer A, Andritsos N, Karabelas AJ, Hoek EMV, Schneider R, Nystrom M (2005) Fouling in nanofiltration. In: Schaefer A, Fane AG, Waite TD (eds) Nanofiltration: principles and applications. Elsevier, Oxford, pp 169–240
Schuch R, Fischetti VA (2009) The secret life of the anthrax agent Bacillus anthracis: bacteriophage-mediated ecological adaptations. PLoS One 4(8):6532
Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR (2006) The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277
Shrout JD, Tolker-Nielsen T, Givskov M, Parsek MR (2011) The contribution of cell–cell signaling and motility to bacterial biofilm formation. MRS Bull 36:367–373
Sillankorva S, Oliveira DR, Vieira MJ, Sutherland IW, Azeredo J (2004) Bacteriophage V S1 infection of Pseudomonas fluorescens planktonic cells versus biofilms. Biofouling 20:133–138
Simões M, Simões LC, Vieira MJ (2010) A review of current and emergent biofilm control strategies. LWT-Food Sci Technol 43:573–583
Siringan P, Connerton PL, Payne RJH, Connerton IF (2011) Bacteriophage-mediated dispersal of Campylobacter jejuni biofilms. Appl Environ Microbiol 77:3320–3326
Soni KA, NannapanenI R (2010) Removal of Listeria monocytogenes biofilms with bacteriophage. J Food Prot 73:1519–1524
Stepnowski P, Siedlecka EM, Behrend P, Jastorff B (2002) Enhanced photo-degradation of contaminants in petroleum refinery wastewater. Water Res 36(9):2167–2172
Stoodley P, Sauer K, Davies DG, Costerson JW (2002) Biofilms as a complex differentiated community. Annu Rev Microbiol 56:187–209
Sun WJ, Liu CF, Yu L, Cui FJ, Zhou Q, Yu SL, Sun L (2012) A novel bacteriophage KSL-1 of 2-Keto-gluconic acid producer Pseudomonas fluorescens K1005: isolation, characterization and its remedial action. BMC Microbiol 29(12):127
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tanji Y, Hattori K, Suzuki K, Miyanaga K (2008) Spontaneous deletion of a 209-kilobase-pair fragment from the Escherichia coli genome occurs with acquisition of resistance to an assortment of infectious phages. Appl Environ Microbiol 74:4256–4263
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Torbeck L, Raccasi D, Guilfoyle DE, Friedman RL, Hussong D (2011) Burkholderia cepacia: this decision is overdue. PDA J Pharm Sci Tech 65:535–543
Toutain CM, Caizza NC, Zegans ME, O’Toole GA (2007) Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res Microbiol 158:471–477
Vatanyoopaisarn S, Nazli A, Dodd CE, Rees CE, Waites WM (2000) Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl Environ Microbiol 66:860–863
Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J (2008) Living on a surface: swarming and biofilm formation. Trends Microbiol 16:496–506
Vongphayloth K, Rattanavong S, Moore CE, Phetsouvanh R, Wuthiekanun V, Sengdouangphachanh A, Phouminh P, Newton PN, Buisson Y (2012) Burkholderia pseudomallei detection in surface water in Southern Laos using Moore’s swabs. Am J Trop Med Hyg 86:872–877
Wagner M, Loy A (2002) Bacterial community composition and function in sewage treatment systems. Curr Opin Biotechnol 13:218–227
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Willians JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:7213–7218
Wimpenny JWT (2000) An overview of biofilms as functional communities. Cambridge University Press, Cambridge
Xiong Y, Liu Y (2010) Biological control of microbial attachment: a promising alternative for mitigating membrane biofouling. Appl Microbiol Biotechnol 86:825–837
Zhang M, Jiang S, Tanuwidjaja D, Voutchkov N, Hoek EMV, Cai B (2011) Composition and variability of biofouling organisms in seawater reverse osmosis desalination plants. Appl Environ Microbiol 77(13):4390
Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han S (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Belgini, D.R.B., Dias, R.S., Siqueira, V.M. et al. Culturable bacterial diversity from a feed water of a reverse osmosis system, evaluation of biofilm formation and biocontrol using phages. World J Microbiol Biotechnol 30, 2689–2700 (2014). https://doi.org/10.1007/s11274-014-1693-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-014-1693-1