Abstract
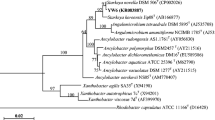
Among the mixture of carbon compounds, bacteria have the ability to select energetically efficient substrate as a preferred carbon source. This specific behavior often limits the use of pollutants as a secondary carbon source by bacteria grown with preferred carbon source and the pollutants. Therefore, to develop an efficient bioremediation technology for removal/degradation of organophosphorus pesticide methyl parathion (MP) from agricultural soils, a soil bacterial strain (LMGR1) capable of degrading MP and p-nitrophenol was isolated and identified as a Cupriavidus oxalaticus based on biochemical (carbon utilization pattern) and molecular analysis (16S rRNA gene sequence), but the phylogenetic analysis revealed a distinct position, and the name was designated as Cupriavidus sp. LMGR1. The effect of preferred carbon source on degradation was also investigated. The bacterium assimilated the pesticide as a sole source of carbon and consumes about 100 mg L−1 of MP within 6 to 7 h of incubation. Furthermore, the methyl parathion hydrolase (MPH) enzyme (localized in the periplasm of bacterium) activity was induced by the carbon deficiency instead of substrate. Determination of kinetic parameters, i.e., Vmax and Km for MPH enzyme in the extracted periplasmic protein indicated significant higher affinity towards the substrate. The reduced MPH activity in the bacterium grown in presence of preferred carbon source indicated the role of carbon catabolite repression in regulation of pesticide degradation. Thus, the bacterium has prodigious capability to be used as a bioremediation agent, but the availability of preferred carbon source may affect its potential in field application.










Similar content being viewed by others
References
Chaudhry, G. R., Ali, A. N., & Wheeler, W. B. (1988). Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the opd gene from a Flavobacterium sp. Applied and Environmental Microbiology, 54, 288–293.
Choudhary, A., Modak, A., Apte, S. K., & Phale, P. S. (2017). Transcriptional modulation of transport-and metabolism-associated gene clusters leading to utilization of benzoate in reference to glucose in Pseudomonas putida CSV86. Applied and Environmental Microbiology, 83, 1280–1217.
Cuadrado, V., Gomila, M., Merini, L., Giulietti, A. M., & Moore, E. R. (2010). Cupriavidus pampae sp. nov., a novel herbicide-degrading bacterium isolated from agricultural soil. International Journal of Systematic and Evolutionary Microbiology, 60, 2606–2612.
de Queiroz, V. T., Azevedo, M. M., da Silva Quadros, I. P., Costa, A. V., do Amaral, A. A., Juvanhol, R. S., et al. (2018). Environmental risk assessment for sustainable pesticide use in coffee production. Journal of Contaminant Hydrology, 219, 18–27.
Deng, S., Chen, Y., Wang, D., Shi, T., Wu, X., Ma, X., et al. (2015). Rapid biodegradation of organophosphorus pesticides by Stenotrophomonas sp. G1. Journal of Hazardous Materials, 297, 17–24.
Donoso, R. A., Pérez-Pantoja, D., & González, B. (2011). Strict and direct transcriptional repression of the pobA gene by benzoate avoids 4-hydroxybenzoate degradation in the pollutant degrader bacterium Cupriavidus necator JMP134. Environmental Microbiology, 13, 1590–1600.
Görke, B., & Stülke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nature Reviews. Microbiology, 6, 613.
Heuer, H., Krsek, M., Baker, P., Smalla, K., & Wellington, E. M. H. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology, 63, 3233–3241.
Holmes, Á., Costas, Á., Ganner, Á., On, S. L., & Stevens, M. (1994). Evaluation of biolog system for identification of some gram-negative bacteria of clinical importance. Journal of Clinical Microbiology, 32, 1970–1975.
Horemans, B., Breugelmans, P., Hofkens, J., & Springael, D. (2017). Carbon catabolite repression and cell dispersal affect degradation of the xenobiotic compound 3, 4-dichloroaniline in Comamonas testosteroni WDL7 biofilms. FEMS Microbiology Ecology, 93, 1–8.
Kegley SE, Hill BR, Orme S, Choi AH (2014) PAN pesticide Database, PesticideAction Network, North America. http://www.pesticideinfo.org (accessed 14November 2016).
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
Kumar, S., Kaushik, G., Dar, M. A., Nimesh, S., Lopez-Chuken, U. J., & Villarreal-Chiu, J. F. (2018). Microbial degradation of organophosphate pesticides: a review. Pedosphere, 28, 190–208.
Lu, P., Li, Q., Liu, H., Feng, Z., Yan, X., Hong, Q., & Li, S. (2013). Biodegradation of chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol by Cupriavidus sp. DT-1. Bioresource Technology, 127, 337–342.
Mitra, A., Chatterjee, C., & Mandal, F. B. (2011). Synthetic chemical pesticides and their effects on birds. Res. J. Environmental Toxicology, 5, 81–96.
Neumann, B., Pospiech, A., & Schairrer, H. U. (1992). Rapid isolationof genomic DNA from gram-negative bacteria. Trends in Genetics, 8, 332–333.
Patnaik, R., & Padhy, R. N. (2016). Evaluation of geno-toxicity of methyl parathion and chlorpyrifos to human liver carcinoma cell line (HepG2). Environmental Science and Pollution Research, 23, 8492–8499.
Perez-Pantoja, D., De la Iglesia, R., Pieper, D. H., & González, B. (2008). Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiology Reviews, 32, 736–794.
Ragnarsdottir, K. V. (2000). Environmental fate and toxicology of organophosphate pesticides. J Geological Soc, 157, 859–876.
Rani, N. L., & Lalithakumari, D. (1994). Degradation of methyl parathion by Pseudomonas putida. Canadian Journal of Microbiology, 40, 1000–1006.
Rayu, S., Nielsen, U. N., Nazaries, L., & Singh, B. K. (2017). Isolation and molecular characterization of novel chlorpyrifos and 3,5,6-trichloro-2-pyridinol-degrading bacteria from sugarcane farm soils. Frontiers in Microbiology, 8, 518.
Sahin, N., Isik, K., Tamer, A. U., & Goodfellow, M. (2000). Taxonomic position of “Pseudomonas oxalaticus” strain Ox1T (DSM 1105T) (Khambata and Bhat, 1953) and its description in the genus Ralstonia as Ralstonia oxalatica comb. nov. Systematic and Applied Microbiology, 23, 206–209.
Sharma, J. (2015). A review on in situ biodegradation of methyl parathion through soilmicrobes. International Journal of Current Microbiology and Applied Sciences, 4, 632–649.
Singh, B. K. (2009). Organophosphorus-degrading bacteria: ecology and industrial applications. Nature Reviews. Microbiology, 7, 156–164.
Singh, B. K., & Walker, A. (2006). Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews, 3, 428–471.
Spain, J. C., & Gibson, D. T. (1991). Pathway for biodegradation of p-Nitrophenol in a Moraxella sp. Applied and Environmental Microbiology, 57, 812–819.
Vandamme, P., & Coenye, T. (2004). Taxonomy of the genus Cupriavidus: a tale of lost and found. International Journal of Systematic and Evolutionary Microbiology, 54, 2285–2289.
Wang, S. (2007). Isolation and identification of methyl parathion degrading bacteria and cloning of hydrolase gene. China: Shandong Agricultural University.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., & Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology, 173, 697–703.
Xu W (2005) Purification and characterization of methyl parathion hydrolase from Plesiomonas sp. M6, Nanjing Agricultural University China.
Yang, L., Zhao, Y. H., Zhang, B. X., Yang, C. H., & Zhang, X. (2005). Isolation and characterization of a chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol degrading bacterium. FEMS Microbiology Letters, 251, 67–73.
Yang, C., Liu, N., Guo, X., & Qiao, C. (2006). Cloning of mpd gene from a chlorpyrifos-degrading bacterium and use of this strain in bioremediation of contaminated soil. FEMS Microbiology Letters, 265, 118–125.
Zhang, R., Cui, Z., Jiang, J., He, J., Gu, X., & Li, S. (2005). Diversity of organophosphorus pesticide-degrading bacteria in a polluted soil and conservation of their organophosphorus hydrolase genes. Canadian Journal of Microbiology, 51, 337–343.
Zhongli, C., Shunpeng, L., & Guoping, F. (2001). Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Applied and Environmental Microbiology, 67, 4922–4925.
Acknowledgment
We are thankful to the Head, Department of Botany, Banaras Hindu University, Varanasi, India, for providing necessary laboratory facilities. Thanks are also due to AIRF, JNU, New Delhi, India, for GCMS analysis and NCMR-NCCS, Pune, India for 16S rRNA gene sequencing. Balkrishna Tiwari is also thankful to the ICAR-AMAAS for financial help in the form of SRF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tiwari, B., Sindhu, V., Mishra, A.K. et al. Carbon Catabolite Repression of Methyl Parathion Degradation in a Bacterial Isolate Characterized as a Cupriavidus sp. LMGR1. Water Air Soil Pollut 231, 357 (2020). https://doi.org/10.1007/s11270-020-04715-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11270-020-04715-7