Abstract
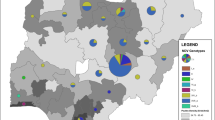
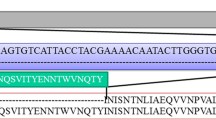
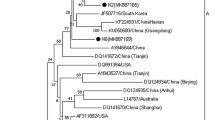
The virus that causes Marek’s disease (MD) is globally ubiquitous in chickens, continuously evolving, and poses a significant threat to the poultry industry. Although vaccines are extensively used, MD still occurs frequently and the virus has evolved increased virulence in China. Here, we report an outbreak of MD in vaccinated chickens and unvaccinated turkeys in a backyard farm in Guangdong province, China, in 2018. Phylogenetic analysis revealed two lineages of MDVs at this farm, with one lineage, containing isolates from two turkeys and five chickens, clustering with virulent Chinese strains and displays a relatively high genetic divergence from the vaccine strains. These new isolates appear to have broken through vaccine immunity, yielding this outbreak of MD in chickens and turkeys. The second lineage included four chicken isolates that clustered with the CVI988 and 814 vaccine strains. The large diversity of MDVs in this single outbreak reveals a complex circulation of MDVs in China. Poor breeding conditions and the weak application of disease prevention and control measures make backyard farms a hotbed for the evolution of viruses that cause infectious diseases. This is especially important in MDV as the MD vaccines do not provide sterilizing immunity, which allows the replication and shedding of virulent field viruses by vaccinated individuals and supporting the continuous evolution of MDVs. Hence, constant monitoring of the evolution of MDVs is necessary to understand the evolution of these field viruses and potential expansions of their host range.



Similar content being viewed by others
Data availability
The sequences generated in the current study were deposited to the GenBank under the accession numbers OP887017-OP887027.
Abbreviations
- MD:
-
Marek’s disease
- MDV:
-
Marek’s disease virus
- HVT:
-
Turkey herpesvirus
References
Calnek BW (2001) Pathogenesis of Marek’s disease virus infection. Curr Top Microbiol Immunol 255:25–55. https://doi.org/10.1007/978-3-642-56863-3_2
Davison AJ, Eberle R, Ehlers B et al (2009) The order Herpesvirales. Arch Virol 154:171–177. https://doi.org/10.1007/s00705-008-0278-4
Witter RL (1983) Characteristics of Marek’s disease viruses isolated from vaccinated commercial chicken flocks: association of viral pathotype with lymphoma frequency. Avian Dis 27:113–132
Shamblin CE, Greene N, Arumugaswami V et al (2004) Comparative analysis of Marek’s disease virus (MDV) glycoprotein-, lytic antigen pp38- and transformation antigen Meq-encoding genes: association of meq mutations with MDVs of high virulence. Vet Microbiol 102:147–167. https://doi.org/10.1016/j.vetmic.2004.06.007
Liao Y, Lupiani B, Ai-Mahmood M et al (2021) Marek’s disease virus US3 protein kinase phosphorylates chicken HDAC 1 and 2 and regulates viral replication and pathogenesis. PLoS Pathog 17:e1009307. https://doi.org/10.1371/journal.ppat.1009307
Osterrieder N, Kamil JP, Schumacher D et al (2006) Marek’s disease virus: from miasma to model. Nat Rev Microbiol 4:283–294. https://doi.org/10.1038/nrmicro1382
Nair V (2018) Spotlight on avian pathology: Marek’s disease. Avian Pathol 47:440–442. https://doi.org/10.1080/03079457.2018.1484073
Zhuang X, Zou H, Shi H et al (2015) Outbreak of Marek’s disease in a vaccinated broiler breeding flock during its peak egg-laying period in China. BMC Vet Res 11:157. https://doi.org/10.1186/s12917-015-0493-7
Baigent SJ, Smith LP, Nair VK et al (2006) Vaccinal control of Marek’s disease: current challenges, and future strategies to maximize protection. Vet Immunol Immunopathol 112:78–86. https://doi.org/10.1016/j.vetimm.2006.03.014
Reddy SM, Izumiya Y, Lupiani B (2017) Marek’s disease vaccines: current status, and strategies for improvement and development of vector vaccines. Vet Microbiol 206:113–120. https://doi.org/10.1016/j.vetmic.2016.11.024
Sun A, Zhao X, Zhu X et al (2022) Fully attenuated meq and pp38 double gene deletion mutant virus confers superior immunological protection against highly virulent Marek’s disease virus infection. Microbiol Spectr 10:e0287122. https://doi.org/10.1128/spectrum.02871-22
Gimeno IM, Witter RL, Hunt HD et al (2005) The pp38 gene of Marek’s disease virus (MDV) is necessary for cytolytic infection of B cells and maintenance of the transformed state but not for cytolytic infection of the feather follicle epithelium and horizontal spread of MDV. J Virol 79:4545–4549. https://doi.org/10.1128/jvi.79.7.4545-4549.2005
Lupiani B, Lee LF, Cui X et al (2004) Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc Natl Acad Sci USA 101:11815–11820. https://doi.org/10.1073/pnas.0404508101
Shi M, Li M, Wang P et al (2021) An outbreak in three-yellow chickens with clinical tumors of high mortality caused by the coinfection of reticuloendotheliosis virus and Marek’s disease virus: a speculated reticuloendotheliosis virus contamination plays an important role in the case. Poult Sci 100:19–25. https://doi.org/10.1016/j.psj.2020.09.034
Chen S, Zhou Y, Chen Y et al (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Li H, Durbin R (2010) Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26:589–595. https://doi.org/10.1093/bioinformatics/btp698
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Kurtz S, Phillippy A, Delcher AL et al (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12. https://doi.org/10.1186/gb-2004-5-2-r12
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Nguyen LT, Schmidt HA, von Haeseler A et al (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Martin DP, Murrell B, Golden M et al (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. https://doi.org/10.1093/ve/vev003
Lole KS, Bollinger RC, Paranjape RS et al (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. https://doi.org/10.1128/jvi.73.1.152-160.1999
Davidson I, Malkinson M, Weisman Y (2002) Marek’s disease in turkeys. I. A seven-year survey of commercial flocks and experimental infection using two field isolates. Avian Dis 46:314–321. https://doi.org/10.1637/0005-2086(2002)046[0314:Msditi]2.0.Co;2
Mescolini G, Lupini C, Davidson I et al (2020) Molecular characterization of a Marek’s disease virus strain detected in tumour-bearing turkeys. Avian Pathol 49:202–207. https://doi.org/10.1080/03079457.2019.1691715
Kenzy SG, Cho BR (1969) Transmission of classical Marek’s disease by affected and carrier birds. Avian Dis 13:211–214
Haesendonck R, Garmyn A, Dorrestein GM et al (2015) Marek’s disease virus associated ocular lymphoma in Roulroul partridges (Rollulus rouloul). Avian Pathol 44:347–351. https://doi.org/10.1080/03079457.2015.1056088
Murata S, Chang KS, Yamamoto Y et al (2007) Detection of the virulent Marek’s disease virus genome from feather tips of wild geese in Japan and the Far East region of Russia. Arch Virol 152:1523–1526. https://doi.org/10.1007/s00705-007-0982-5
Lian X, Ming X, Xu J et al (2018) First molecular detection and characterization of Marek’s disease virus in red-crowned cranes (Grus japonensis): a case report. BMC Vet Res 14:122. https://doi.org/10.1186/s12917-018-1437-9
Murata S, Hayashi Y, Kato A et al (2012) Surveillance of Marek’s disease virus in migratory and sedentary birds in Hokkaido, Japan. Vet J 192:538–540. https://doi.org/10.1016/j.tvjl.2011.07.006
Murata S, Machida Y, Isezaki M et al (2020) Genetic characterization of a Marek’s disease virus strain isolated in Japan. Virol J 17:186. https://doi.org/10.1186/s12985-020-01456-1
Li Y, Sun A, Su S et al (2011) Deletion of the Meq gene significantly decreases immunosuppression in chickens caused by pathogenic Marek’s disease virus. Virol J 8:2. https://doi.org/10.1186/1743-422x-8-2
Zhang YP, Lv HC, Bao KY et al (2016) Molecular and pathogenicity characterization of Gallid herpesvirus 2 newly isolated in China from 2009 to 2013. Virus Genes 52:51–60. https://doi.org/10.1007/s11262-015-1264-z
Sato J, Murata S, Yang Z et al (2022) Effect of insertion and deletion in the meq protein encoded by highly oncogenic Marek’s disease virus on transactivation activity and virulence. Viruses. https://doi.org/10.3390/v14020382
Murata S, Okada T, Kano R et al (2011) Analysis of transcriptional activities of the Meq proteins present in highly virulent Marek’s disease virus strains, RB1B and Md5. Virus Genes 43:66–71. https://doi.org/10.1007/s11262-011-0612-x
Padhi A, Parcells MS (2016) Positive selection drives rapid evolution of the meq oncogene of Marek’s disease virus. PLoS ONE 11:e0162180. https://doi.org/10.1371/journal.pone.0162180
Trimpert J, Groenke N, Jenckel M et al (2017) A phylogenomic analysis of Marek’s disease virus reveals independent paths to virulence in Eurasia and North America. Evol Appl 10:1091–1101. https://doi.org/10.1111/eva.12515
Yu ZH, Teng M, Luo J et al (2013) Molecular characteristics and evolutionary analysis of field Marek’s disease virus prevalent in vaccinated chicken flocks in recent years in China. Virus Genes 47:282–291. https://doi.org/10.1007/s11262-013-0942-y
Li K, Yu Z, Lan X et al (2022) Complete genome analysis reveals evolutionary history and temporal dynamics of Marek’s disease virus. Front Microbiol 13:1046832. https://doi.org/10.3389/fmicb.2022.1046832
Zhang Y, Lan X, Wang Y et al (2022) Emerging natural recombinant Marek’s disease virus between vaccine and virulence strains and their pathogenicity. Transbound Emerg Dis 69:e1702–e1709. https://doi.org/10.1111/tbed.14506
Cui H, Gao H, Cui X et al (2013) Avirulent Marek’s disease virus type 1 strain 814 vectored vaccine expressing avian influenza (AI) virus H5 haemagglutinin induced better protection than turkey herpesvirus vectored AI vaccine. PLoS ONE 8:e53340. https://doi.org/10.1371/journal.pone.0053340
Reddy SK, Sharma JM, Ahmad J et al (1996) Protective efficacy of a recombinant herpesvirus of turkeys as an in ovo vaccine against Newcastle and Marek’s diseases in specific-pathogen-free chickens. Vaccine 14:469–477. https://doi.org/10.1016/0264-410x(95)00242-s
Sun GR, Zhang YP, Lv HC et al (2017) A chinese variant Marek’s disease virus strain with divergence between virulence and vaccine resistance. Viruses 9:71. https://doi.org/10.3390/v9040071
Baigent SJ, Petherbridge LJ, Smith LP et al (2006) Herpesvirus of turkey reconstituted from bacterial artificial chromosome clones induces protection against Marek’s disease. J Gen Virol 87:769–776. https://doi.org/10.1099/vir.0.81498-0
Acknowledgements
This work was supported by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007), Laboratory of Lingnan Modern Agriculture Project (NT2021007), the Guangdong Provincial Key R&D Program, (2022B1111040001), the National Natural Science Foundation of China (31822056), the Guangdong Science and Technology Innovation Leading Talent Program (2019TX05N098), the 111 Project (D20008), the Double first-class discipline promotion project (2023B10564003) and the Department of Education of Guangdong Province (2019KZDXM004 and 2019KCXTD001).
Funding
This study was funded by Guangdong Major Project of Basic and Applied Basic Research, 2020B0301030007.
Author information
Authors and Affiliations
Contributions
YS conceptualized, designed and and wrote the manuscript. WL, HM, JP and XS performed the experiments. XL analyzed the data. DMI participated in revising the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Edited by Juergen Richt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, W., Meng, H., Liang, X. et al. The genome evolution of Marek’s disease viruses in chickens and turkeys in China. Virus Genes 59, 845–851 (2023). https://doi.org/10.1007/s11262-023-02034-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-023-02034-7