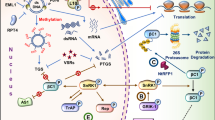
Abstract
Viruses belonging to the family Geminiviridae infect plants and are responsible for a number of diseases of crops in the tropical and sub-tropical regions of the World. The innate immune response of the plant assists in its defense against such viral pathogens by the recognition of pathogen/microbe-associated molecular patterns through pattern-recognition receptors. Phytohormone signalling pathways play a vital role in plant defense responses against these devastating viruses. Geminiviruses, however, have developed counter-defense strategies that prevail over the above defense pathways. The proteins encoded by geminiviruses act as suppressors of plant immunity by interacting with the signalling components of several hormones. In this review we focus on the molecular interplay of phytohormone pathways and geminiviral infection and try to find interesting parallels with similar mechanisms known in other plant-infecting viruses and strengthen the argument that this interplay is necessary for disease development.



Similar content being viewed by others
Data availability
The authors declare that no data has been generated in this manuscript and hence there is no data to be made available.
Code availability
The authors declare that no specialized software has been used to prepare this manuscript and hence there is no code to be made available.
References
Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad spectrum disease resistance. Annu Rev Phytopathol 55:257–286
Ranf S (2017) Sensing of molecular patterns through cell surface immune receptors. Curr Opin Plant Biol 38:68–77. https://doi.org/10.1016/j.pbi.2017.04.011
Saijo Y, E. P. IIan Loo, and S. Yasuda, (2018) Pattern recognition receptors and signaling in plant–microbe interactions. Plant J 93(4):592–613. https://doi.org/10.1111/tpj.13808
Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8(4):521–539. https://doi.org/10.1016/j.molp.2014.12.022
Thulasi Devendrakumar K, Li X, Zhang Y (2018) MAP kinase signalling: interplays between plant PAMP- and effector-triggered immunity. Cell Mol Life Sci 75(16):2981–2989. https://doi.org/10.1007/s00018-018-2839-3
Nicaise V, Candresse T (2017) Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol Plant Pathol 18(6):878–886. https://doi.org/10.1111/mpp.12447
Kong J et al (2018) The cucumber mosaic virus movement protein suppresses PAMP-triggered immune responses in Arabidopsis and tobacco. Biochem Biophys Res Commun 498(3):395–401. https://doi.org/10.1016/j.bbrc.2018.01.072
Niehl A, Wyrsch I, Boller T, Heinlein M (2016) Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol 211(3):1008–1019. https://doi.org/10.1111/nph.13944
Amari K, Niehl A (2020) Nucleic acid-mediated PAMP-triggered immunity in plants. Curr Opin Virol 42:32–39. https://doi.org/10.1016/j.coviro.2020.04.003
Jones JDJ, Dangl JL (2006) The plant immune system. Nature 444(7117):323–329
Caplan J, Padmanabhan M, Dinesh-Kumar SP (2008) Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3(3):126–135. https://doi.org/10.1016/j.chom.2008.02.010
Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. https://doi.org/10.1146/annurev-arplant-050213-040012
Bürger M, Chory J (2019) Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe 26(2):163–172. https://doi.org/10.1016/j.chom.2019.07.006
Santner A, Estelle M (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459(7250):1071–1078. https://doi.org/10.1038/nature08122
Pieterse CMJ, Van Der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28(April):489–521. https://doi.org/10.1146/annurev-cellbio-092910-154055
Alazem M, Lin NS (2015) Roles of plant hormones in the regulation of host-virus interactions. Mol Plant Pathol 16(5):529–540. https://doi.org/10.1111/mpp.12204
Ma KW, Ma W (2016) Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol Biol 91(6):713–725. https://doi.org/10.1007/s11103-016-0452-0
Islam W, Naveed H, Zaynab M, Huang Z, Chen HYH (2019) Plant defense against virus diseases; growth hormones in highlights. Plant Signal Behav 14(6):1–10. https://doi.org/10.1080/15592324.2019.1596719
Rojas MR et al (2018) World Management of Geminiviruses. Annu Rev Phytopathol 56:637–677. https://doi.org/10.1146/annurev-phyto-080615-100327
Varsani A et al (2014) Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 159(8):2193–2203. https://doi.org/10.1007/s00705-014-2050-2
Zerbini FM et al (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98(2):131–133. https://doi.org/10.1099/jgv.0.000738
Gnanasekaran P, KishoreKumar R, Bhattacharyya D, Vinoth Kumar R, Chakraborty S (2019) Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol Plant Pathol 20(7):1019–1033. https://doi.org/10.1111/mpp.12800
Van Wezel R, Dong X, Blake P, Stanley J, Hong Y (2002) Differential roles of geminivirus Rep and AC4 (C4) in the induction of necrosis in Nicotiana benthamiana. Mol Plant Pathol 3(6):461–471. https://doi.org/10.1046/j.1364-3703.2002.00141.x
Hussain M, Mansoor S, Iram S, Zafar Y, Briddon RW (2007) The hypersensitive response to tomato leaf curl New Delhi virus nuclear shuttle protein is inhibited by transcriptional activator protein. Mol Plant-Microbe Interact 20(12):1581–1588. https://doi.org/10.1094/MPMI-20-12-1581
Sharma P, Ikegami M (2010) Tomato leaf curl Java virus V2 protein is a determinant of virulence, hypersensitive response and suppression of posttranscriptional gene silencing. Virology 396(1):85–93. https://doi.org/10.1016/j.virol.2009.10.012
Saeed F et al (2018) Infectivity of okra enation leaf curl virus and the role of its V2 protein in pathogenicity. Virus Res 255(January):90–94. https://doi.org/10.1016/j.virusres.2018.07.007
Shine MB, Xiao X, Kachroo P, Kachroo A (2019) Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci 279:81–86. https://doi.org/10.1016/j.plantsci.2018.01.001
Ghosh D, Chakraborty S (2021) Molecular interplay between phytohormones and geminiviruses: a saga of a never-ending arms race. J Exp Bot 72(8):2903–2917. https://doi.org/10.1093/jxb/erab061
Gupta N, Reddy K, Bhattacharyya D, Chakraborty S (2021) Plant responses to geminivirus infection: guardians of the plant immunity. Virol J 18(1):1–25. https://doi.org/10.1186/s12985-021-01612-1
Qi G et al (2018) Pandemonium Breaks Out: Disruption of Salicylic Acid-Mediated Defense by Plant Pathogens. Mol Plant 11(12):1427–1439. https://doi.org/10.1016/j.molp.2018.10.002
Janda T, Szalai G, Pál M (2020) Salicylic acid signalling in plants. Int J Mol Sci. https://doi.org/10.3390/ijms21072655
Ding P, Ding Y (2020) Stories of salicylic acid: a plant defense hormone. Trends Plant Sci 25(6):549–565. https://doi.org/10.1016/j.tplants.2020.01.004
Lefevere H, Bauters L, Gheysen G (2020) Salicylic Acid Biosynthesis in Plants. Front Plant Sci 11(April):1–7. https://doi.org/10.3389/fpls.2020.00338
Tada Y et al (2008) Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321(5891):952–956. https://doi.org/10.1126/science.1156970
Zhou JM et al (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant-Microbe Interact 13(2):191–202. https://doi.org/10.1094/MPMI.2000.13.2.191
Li N, Han X, Feng D, Yuan D, Huang LJ (2019) Signaling crosstalk between salicylic acid and ethylene/Jasmonate in plant defense: do we understand what they are whispering? Int J Mol Sci. https://doi.org/10.3390/ijms20030671
Pokotylo I, Kravets V, Ruelland E (2019) Salicylic acid binding proteins (SABPs): the hidden forefront of salicylic acid signalling. Int J Mol Sci 20(18):1–20. https://doi.org/10.3390/ijms20184377
Alamillo JM, Saénz P, García JA (2006) Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J 48(2):217–227. https://doi.org/10.1111/j.1365-313X.2006.02861.x
Agudelo-Romero P et al (2008) Changes in the gene expression profile of Arabidopsis thaliana after infection with tobacco etch virus. Virol J 5:1–11. https://doi.org/10.1186/1743-422X-5-92
Chen H et al (2010) Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J 62(1):12–23. https://doi.org/10.1111/j.1365-313X.2009.04120.x
Rodriguez MC, Conti G, Zavallo D, Manacorda CA, Asurmendi S (2014) TMV-Cg Coat Protein stabilizes DELLA proteins and in turn negatively modulates salicylic acid-mediated defense pathway during Arabidopsis thaliana viral infection. BMC Plant Biol 14(1):1–17. https://doi.org/10.1186/s12870-014-0210-x
Tian M et al (2015) Salicylic acid inhibits the replication of tomato bushy stunt virus by directly targeting a host component in the replication complex. Mol Plant-Microbe Interact 28(4):379–386. https://doi.org/10.1094/MPMI-09-14-0259-R
K. Li et al., 2018 “Transcriptome analysis of Nicotiana benthamiana infected by Tobacco curly shoot virus,” Viro. J Doi: https://doi.org/10.1186/s12985-018-1044-1
Wang D, Zhang X, Yao X, Zhang P, Fang R, Ye J (2020) A 7-amino-acid motif of REp protein essential for virulence is critical for triggering host defense against Sri Lankan cassava mosaic virus. Mol Plant-Microbe Interact 33(1):78–86. https://doi.org/10.1094/MPMI-06-19-0163-FI
Ascencio-Ibáñez JT et al (2008) Global analysis of arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148(1):436–454. https://doi.org/10.1104/pp.108.121038
Miozzi L, Napoli C, Sardo L, Accotto GP (2014) Transcriptomics of the interaction between the monopartite phloem-limited geminivirus tomato yellow leaf curl sardinia virus and solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during infection. PLoS ONE. https://doi.org/10.1371/journal.pone.0089951
Yang LP et al (2013) C2-mediated decrease in DNA methylation, accumulation of siRNAs, and increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus. Plant J 73(6):910–917. https://doi.org/10.1111/tpj.12081
Tu YC et al (2017) The C2 protein of tomato leaf curl Taiwan virus is a pathogenicity determinant that interferes with expression of host genes encoding chromomethylases. Physiol Plant 161(4):515–531. https://doi.org/10.1111/ppl.12615
Guerrero J, Regedanz E, Lu L, Ruan J, Bisaro DM, Sunter G (2020) Manipulation of the plant host by the Geminivirus AC2/C2 protein, a central player in the infection cycle. Front Plant Sci 11(May):1–18. https://doi.org/10.3389/fpls.2020.00591
Matić S, Pegoraro M, Noris E (2016) The C2 protein of tomato yellow leaf curl Sardinia virus acts as a pathogenicity determinant and a 16-amino acid domain is responsible for inducing a hypersensitive response in plants. Virus Res 215:12–19. https://doi.org/10.1016/j.virusres.2016.01.014
Shi X et al (2013) Plant virus differentially alters the plant’s defense response to its closely related vectors. PLoS ONE 8(12):1–8. https://doi.org/10.1371/journal.pone.0083520
C. Hettenhausen, M. C. Schuman, and J. Wu, “Europe PMC Funders Group Europe PMC Funders Author Manuscripts MAPK signaling – a key element in plant defense response to insects,” vol. 22, no. 2, pp. 157–164, 2017, doi: https://doi.org/10.1111/1744-7917.12128.MAPK
Yu G et al (2020) A bacterial effector protein prevents mapk-mediated phosphorylation of sgt1 to suppress plant immunity. PLoS Pathog 16(9):1–30. https://doi.org/10.1371/journal.ppat.1008933
Li Y et al (2017) SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum). PLoS ONE 12(2):1–21. https://doi.org/10.1371/journal.pone.0172466
Zhang L, Zhang F, Melotto M, Yao J, He SY (2017) Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot 68(6):1371–1385. https://doi.org/10.1093/jxb/erw478
Wasternack C, Hause B (2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111(6):1021–1058. https://doi.org/10.1093/aob/mct067
Fonseca S et al (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5(5):344–350. https://doi.org/10.1038/nchembio.161
J. Ruan et al., 2019 “Jasmonic acid signaling pathway in plants,” Int J Mo. Sci, doi: https://doi.org/10.3390/ijms20102479
M. S. Ali and K. H. Baek, 2020 “Jasmonic acid signaling pathway in response to abiotic stresses in plants,” Int. J Mol Sci doi: https://doi.org/10.3390/ijms21020621
Thines B et al (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448(7154):661–665. https://doi.org/10.1038/nature05960
Katsir L, Schilmiller AL, Staswick PE, Sheng YH, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A 105(19):7100–7105. https://doi.org/10.1073/pnas.0802332105
L. B. Sheard et al., 2011 “HHS Public Access,” vol. 468, no. 7322, pp. 400–405, , doi: https://doi.org/10.1038/nature09430.Jasmonate.
Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6(3):686–703. https://doi.org/10.1093/mp/sss128
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10(February):1–17. https://doi.org/10.3389/fpls.2019.00228
Wu X, Ye J (2020) Manipulation of jasmonate signaling by plant viruses and their insect vectors. Viruses 12(2):1–16. https://doi.org/10.3390/v12020148
Góngora-Castillo E, Ibarra-Laclette E, Trejo-Saavedra DL, Rivera-Bustamante RF (2012) Transcriptome analysis of symptomatic and recovered leaves of geminivirus-infected pepper (Capsicum annuum). Virol J 9:1–16. https://doi.org/10.1186/1743-422X-9-295
Csorba T, Kontra L, Burgyán J (2015) Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 479–480:85–103. https://doi.org/10.1016/j.virol.2015.02.028
Lozano-Durán R et al (2011) Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23(3):1014–1032. https://doi.org/10.1105/tpc.110.080267
Li T, Huang Y, Xu ZS, Wang F, Xiong AS (2019) Salicylic acid-induced differential resistance to the tomato yellow leaf curl virus among resistant and susceptible tomato cultivars. BMC Plant Biol 19(1):1–14. https://doi.org/10.1186/s12870-019-1784-0
Jia Q et al (2016) CLCuMuB βC1 Subverts Ubiquitination by Interacting with NbSKP1s to Enhance Geminivirus Infection in Nicotiana benthamiana. PLoS Pathog 12(6):1–30. https://doi.org/10.1371/journal.ppat.1005668
Zou C et al (2020) Begomovirus-associated betasatellite virulence factor βC1 attenuates tobacco defense to whiteflies via interacting with plant SKP1. Front Plant Sci 11(August):1–11. https://doi.org/10.3389/fpls.2020.574557
Li Y et al (2019) Interaction between Brassica yellows virus silencing suppressor P0 and plant SKP1 facilitates stability of P0 in vivo against degradation by proteasome and autophagy pathways. New Phytol 222(3):1458–1473. https://doi.org/10.1111/nph.15702
Chen S et al (2019) Cucurbit chlorotic yellows virus p22 protein interacts with cucumber SKP1LB1 and its F-box-like motif is crucial for silencing suppressor activity. Viruses. https://doi.org/10.3390/v11090818
He L et al (2020) Rice black-streaked dwarf virus-encoded P5–1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol 225(2):896–912. https://doi.org/10.1111/nph.16066
Wu D et al (2017) Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res 27(3):402–415. https://doi.org/10.1038/cr.2017.2
De Geyter N, Gholami A, Goormachtig S, Goossens A (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17(6):349–359. https://doi.org/10.1016/j.tplants.2012.03.001
Wasternack C, Strnad M (2019) Jasmonates are signals in the biosynthesis of secondary metabolites — Pathways, transcription factors and applied aspects — A brief review. N Biotechnol 48(September):1–11. https://doi.org/10.1016/j.nbt.2017.09.007
Luan JB et al (2013) Suppression of terpenoid synthesis in plants by a virus promotes its mutualism with vectors. Ecol Lett 16(3):390–398. https://doi.org/10.1111/ele.12055
Li R et al (2014) Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26(12):4991–5008. https://doi.org/10.1105/tpc.114.133181
Rosas-Díaz T, Macho AP, Beuzón CR, Lozano-Durán R, Bejarano ER (2016) The C2 protein from the geminivirus tomato yellow leaf curl sardinia virus decreases sensitivity to jasmonates and suppresses jasmonate-mediated defences. Plants 5(1):777–788. https://doi.org/10.3390/plants5010008
Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH (2008) βCl, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22(18):2564–2577. https://doi.org/10.1101/gad.1682208
Sun YC, Pan LL, Ying FZ, Li P, Wang XW, Liu SS (2017) Jasmonic acid-related resistance in tomato mediates interactions between whitefly and whitefly-transmitted virus. Sci Rep 7(1):1–7. https://doi.org/10.1038/s41598-017-00692-w
Du J et al (2020) NSs, the silencing suppressor of tomato spotted wilt orthotospovirus, interferes with JA-regulated host terpenoids expression to attract Frankliniella occidentalis. Front Microbiol 11(December):1–12. https://doi.org/10.3389/fmicb.2020.590451
Dubois M, Van den Broeck L, Inzé D (2018) The pivotal role of ethylene in plant growth. Trends Plant Sci 23(4):311–323. https://doi.org/10.1016/j.tplants.2018.01.003
Kazan K (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20(4):219–229. https://doi.org/10.1016/j.tplants.2015.02.001
Binder BM (2020) Ethylene signaling in plants. J Biol Chem 295(22):7710–7725. https://doi.org/10.1074/jbc.REV120.010854
Ju C, Chang C (2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169(1):85–95. https://doi.org/10.1104/pp.15.00845
Bak A, Patton MKF, Perilla-Henao LM, Aegerter BJ, Casteel CL (2019) Ethylene signaling mediates potyvirus spread by aphid vectors. Oecologia 190(1):139–148. https://doi.org/10.1007/s00442-019-04405-0
Broekgaarden C, Caarls L, Vos IA, Pieterse CMJ, Van Wees SCM (2015) Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol 169(4):2371–2379. https://doi.org/10.1104/pp.15.01020
Soitamo AJ, Jada B, Lehto K (2012) Expression of geminiviral AC2 RNA silencing suppressor changes sugar and jasmonate responsive gene expression in transgenic tobacco plants. BMC Plant Biol 12(1):1. https://doi.org/10.1186/1471-2229-12-204
Chen T et al (2013) Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE 8(11):4–6. https://doi.org/10.1371/journal.pone.0080816
Wu M, Ding X, Fu X, Lozano-Duran R (2019) Transcriptional reprogramming caused by the geminivirus tomato yellow leaf curl virus in local or systemic infections in Nicotiana benthamiana. BMC Genomics 20(1):1–17. https://doi.org/10.1186/s12864-019-5842-7
Song S, Qi T, Wasternack C, Xie D (2014) Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol 21:112–119. https://doi.org/10.1016/j.pbi.2014.07.005
Yang DL et al (2008) Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol Plant 1(3):528–537. https://doi.org/10.1093/mp/ssn021
Qi T et al (2014) Arabidopsis DELLA and JAZ proteins bind the WD-Repeat/ bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26(3):1118–1133. https://doi.org/10.1105/tpc.113.121731
Davière JM, Achard P (2013) Gibberellin signaling in plants. Dev 140(6):1147–1151. https://doi.org/10.1242/dev.087650
Bao S, Hua C, Shen L, Yu H (2020) New insights into gibberellin signaling in regulating flowering in Arabidopsis. J Integr Plant Biol 62(1):118–131. https://doi.org/10.1111/jipb.12892
Hofmann NR (2016) A structure for plant-specific transcription factors: the gras domain revealed. Plant Cell 28(5):993–994. https://doi.org/10.1105/tpc.16.00309
Ito T, Okada K, Fukazawa J, Takahashi Y (2018) DELLA-dependent and -independent gibberellin signaling. Plant Signal Behav 13(3):1–3. https://doi.org/10.1080/15592324.2018.1445933
Navarro L et al (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18(9):650–655. https://doi.org/10.1016/j.cub.2008.03.060
Achard P, Genschik P (2009) Releasing the brakes of plant growth: How GAs shutdown della proteins. J Exp Bot 60(4):1085–1092. https://doi.org/10.1093/jxb/ern301
Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160(1):83–92. https://doi.org/10.1104/pp.112.200956
de Vleesschauwer D et al (2012) Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol 158(4):1833–1846. https://doi.org/10.1104/pp.112.193672
Bari R, Jones JDG (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69(4):473–488. https://doi.org/10.1007/s11103-008-9435-0
Zhu S et al (2005) The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol 139(4):1935–1945. https://doi.org/10.1104/pp.105.072306
Lozano-Durán R, Rosas-Díaz T, Luna AP, Bejarano ER (2011) Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS ONE. https://doi.org/10.1371/journal.pone.0022383
Mills-Lujan K, Deom CM (2010) Geminivirus C4 protein alters Arabidopsis development. Protoplasma 239(1–4):95–110. https://doi.org/10.1007/s00709-009-0086-z
Argueso CT et al (2012) Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002448
E. Zürcher and B. Müller, 2016 Cytokinin Synthesis, Signaling, and Function-Advances and New Insights, Elsevier Inc
Kieber JJ, Schaller GE (2018) Cytokinin signaling in plant development. Development 145(4):1–7. https://doi.org/10.1242/dev.149344
Ashihara H, Stasolla C, Fujimura T, Crozier A (2018) Purine salvage in plants. Phytochemistry 147:89–124. https://doi.org/10.1016/j.phytochem.2017.12.008
Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by Geminivirus AL2 and L2 Proteins. J Virol 79(12):7410–7418. https://doi.org/10.1128/jvi.79.12.7410-7418.2005
Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM (2009) Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol 83(10):5005–5013. https://doi.org/10.1128/jvi.01771-08
Baliji S, Lacatus G, Sunter G (2010) The Interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 402(2):238–247. https://doi.org/10.1016/j.virol.2010.03.023
Seo JK et al (2018) Molecular dissection of distinct symptoms induced by tomato chlorosis virus and tomato yellow leaf curl virus based on comparative transcriptome analysis. Virology. https://doi.org/10.1016/j.virol.2018.01.001
Kim EJ, Russinova E (2020) Brassinosteroid signalling. Curr Biol 30(7):R294–R298. https://doi.org/10.1016/j.cub.2020.02.011
Li Z, He Y (2020) Roles of brassinosteroids in plant reproduction. Int J Mol Sci 21(3):1–16. https://doi.org/10.3390/ijms21030872
Youn JH, Kim TW (2015) Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol Plant 8(4):552–565. https://doi.org/10.1016/j.molp.2014.12.006
Sun Y et al (2010) Integration of Brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19(5):765–777. https://doi.org/10.1016/j.devcel.2010.10.010
Planas-Riverola A, Gupta A, Betegoń-Putze I, Bosch N, Ibanḛs M, Cano-Delgado AI (2019) Brassinosteroid signaling in plant development and adaptation to stress. Dev 146(5):1–11. https://doi.org/10.1242/dev.151894
Ortiz-Morea FA, He P, Shan L, Russinova E (2020) It takes two to tango - Molecular links between plant immunity and brassinosteroid signalling. J Cell Sci 133(22):1–11. https://doi.org/10.1242/jcs.246728
Zhang DW, Deng XG, Fu FQ, Lin HH (2015) Induction of plant virus defense response by brassinosteroids and brassinosteroid signaling in Arabidopsis thaliana. Planta 241(4):875–885. https://doi.org/10.1007/s00425-014-2218-8
C. Julie KØrner, et al (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol Plant-Microbe Interact. https://doi.org/10.1094/MPMI-06-13-0179-R
Park J et al (2011) The arabidopsis thaliana homeobox gene ATHB12 is involved in symptom development caused by geminivirus infection. PLoS ONE. https://doi.org/10.1371/journal.pone.0020054
Deom CM, Alabady MS, Yang L (2021) Early transcriptome changes induced by the Geminivirus C4 oncoprotein: setting the stage for oncogenesis. BMC Genomics 22(1):1–19. https://doi.org/10.1186/s12864-021-07455-y
Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11(11):777–788. https://doi.org/10.1038/nrmicro3117
Mills-Lujan K, Andrews DL, Chou CW, Deom CM (2015) The roles of phosphorylation and SHAGGY-like protein kinases in geminivirus C4 protein induced hyperplasia. PLoS ONE 10(3):1–26. https://doi.org/10.1371/journal.pone.0122356
Piroux N, Saunders K, Page A, Stanley J (2007) Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKη, a component of the brassinosteroid signalling pathway. Virology 362(2):428–440. https://doi.org/10.1016/j.virol.2006.12.034
Mei Y et al (2018) Nucleocytoplasmic shuttling of geminivirus C4 protein mediated by phosphorylation and myristoylation is critical for viral pathogenicity. Mol Plant 11(12):1466–1481. https://doi.org/10.1016/j.molp.2018.10.004
Mei Y, Zhang F, Wang M, Li F, Wang Y, Zhou X (2020) Divergent Symptoms caused by geminivirus-encoded C4 proteins correlate with their ability to bind NbSKη. J Virol 94(20):1–12. https://doi.org/10.1128/jvi.01307-20
Clouse SD (2002) Brassinosteroid signal transduction: clarifying the pathway from ligand perception to gene expression. Mol Cell 10(5):973–982. https://doi.org/10.1016/S1097-2765(02)00744-X
Korasick DA, Enders TA, Strader LC (2013) Auxin biosynthesis and storage forms. J Exp Bot 64(9):2541–2555. https://doi.org/10.1093/jxb/ert080
Weijers D, Wagner D (2016) Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67(February):539–574. https://doi.org/10.1146/annurev-arplant-043015-112122
Padmanabhan MS, Kramer SR, Wang X, Culver JN (2008) Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: reprogramming the auxin response pathway to enhance virus infection. J Virol 82(5):2477–2485. https://doi.org/10.1128/jvi.01865-07
Padmanabhan MS, Goregaoker SP, Golem S, Shiferaw H, Culver JN (2005) Interaction of the tobacco mosaic virus replicase protein with the Aux/IAA protein PAP1/IAA26 is associated with disease development. J Virol 79(4):2549–2558. https://doi.org/10.1128/jvi.79.4.2549-2558.2005
Jin L et al (2016) Rice Dwarf Virus P2 protein Hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog 12(9):1–23. https://doi.org/10.1371/journal.ppat.1005847
Zhang H et al (2019) Suppression of auxin signalling promotes rice susceptibility to rice black streaked dwarf virus infection. Mol Plant Pathol 20(8):1093–1104. https://doi.org/10.1111/mpp.12814
Acknowledgements
The authors acknowledge the financial assistance received from J. C. Bose Fellowship, Science and Engineering Research Board, Government of India to ID (SB/S2/JCB-057/2016). KG acknowledges the fellowship from the Department of Science and Technology (DST), Government of India under Innovation in Science Pursuit for Inspired Research (INSPIRE; grant number DST/INSPIRE Fellowship/2015/IF150761) Faculty Research Program Grant under IoE, University of Delhi.
Funding
F.unding was received from J. C. Bose Fellowship, Science and Engineering Research Board, Government of India to ID (SB/S2/JCB-057/2016), Faculty Research Program Grant under IoE, University of Delhi and the Fellowship from the Department of Science and Technology, Government of India under Innovation in Science Pursuit for Inspired Research (INSPIRE; grant number DST/INSPIRE Fellowship/2015/IF150761) to KG.
Author information
Authors and Affiliations
Contributions
KG, RR and ID conceptualized the review; KG and RR performed the data analysis and prepared the original draft of the manuscript; ID edited the manuscript and arranged the funding.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest /competing interests with respect to this manuscript.
Research involving human participants or animals
The authors declare that this research did not involve any human subjects or animals. Hence, the requirement of approvals by ethics committees does not arise.
Informed consent
The authors declare that this research did not involve collecting data of any human subjects or vulnerable population. Hence, the requirement of taking informed consent does not arise.
Additional information
Edited by Seung-Kook Choi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gupta, K., Rishishwar, R. & Dasgupta, I. The interplay of plant hormonal pathways and geminiviral proteins: partners in disease development. Virus Genes 58, 1–14 (2022). https://doi.org/10.1007/s11262-021-01881-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-021-01881-6