Abstract
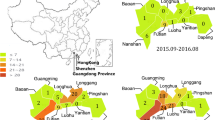
In this study, we investigated the molecular characteristics and spatio-temporal dynamics of GII.P17-GII.17 norovirus in Zhoushan Islands during 2013–2018. We collected 1849 samples from sporadic acute gastroenteritis patients between January 2013 and August 2018 in Zhoushan Islands, China. Among the 1849 samples, 134 (7.24%) samples were positive for human norovirus (HuNoV). The complete sequence of GII.17 VP1 gene was amplified from 31 HuNoV-positive samples and sequenced. A phylogenetic tree was constructed based on the full-length sequence of the VP1 gene. Phylogenetic analysis revealed that the GII.17 genotype detected during 2014–2018 belongs to the new GII.17 Kawasaki variant. Divergence analysis revealed that the time of the most recent common ancestor (TMRCA) of GII.17 in Zhoushan Islands was estimated to be between 1997 and 1998. The evolutionary rate of the VP1 gene of the GII.17 genotype norovirus was 1.14 × 10–3 (95% HPD: 0.62–1.73 × 10–3) nucleotide substitutions/site/year. The spatio-temporal diffusion analysis of the GII.17 genotype identified Hong Kong as the epicenter for GII.17 dissemination. The VP1 gene sequence of Zhoushan Island isolates correlated with that of Hong Kong and Japan isolates.





Similar content being viewed by others
References
Bányai K, Estes MK, Martella V, Parashar UD (2018) Viral gastroenteritis. Lancet 392(10142):175–186
Lopman BA, Steele D, Kirkwood CD, Parashar UD (2016) The vast and varied global burden of norovirus: prospects for prevention and control. PLoS Med 13(4):e1001999
Kapikian AZ, Wyatt RG, Dolin R, Thornhill TS, Kalica AR, Chanock RM (1972) Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol 10(5):1075–1081
Hardy ME (2005) Norovirus protein structure and function. FEMS Microbiol Lett 253(1):1–8
Donaldson EF, Lindesmith LC, Lobue AD, Baric RS (2010) Viral shape-shifting: norovirus evasion of the human immune system. Nat Rev Microbiol 8(3):231–241
Vinjé J (2015) Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 53(2):373–381
Bok K, Green KY (2012) Norovirus gastroenteritis in immunocompromised patients. N Engl J Med 367:2126–2132
Victoria M, Miagostovich MP, Ferreira MS, Vieira CB, Fioretti JM, Leite JP, Colina R, Cristina J (2009) Bayesian coalescent inference reveals high evolutionary rates and expansion of norovirus populations. Infect Genet Evol 9(5):927–932
Parra GI, Squires RB, Karangwa CK, Johnson JA, Lepore CJ, Sosnovtsev SV, Green KY (2017) Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog 13(1):e1006136
Motoya T, Nagasawa K, Matsushima Y, Nagata N, Ryo A, Sekizuka T, Yamashita A, Kuroda M, Morita Y, Suzuki Y, Sasaki N, Katayama K, Kimura H (2017) Molecular evolution of the VP1 gene in human norovirus GII.4 variants in 1974–2015. Front Microbiol 8:2399
Eden JS, Hewitt J, Lim KL, Boni MF, Merif J, Greening G, Ratcliff RM, Holmes EC, Tanaka MM, Rawlinson WD, White PA (2014) The emergence and evolution of the novel epidemic norovirus GII.4 variant Sydney 2012. Virology 450–451:106–113
Ao Y, Cong X, Jin M, Sun X, Wei X, Wang J, Zhang Q, Song J, Yu J, Cui J, Qi J, Tan M, Duan Z (2018) Genetic analysis of re-emerging GIIP.16–GII.2 noroviruses in 2016–2017 in China. J Infect Dis 29:1–33
Yu Y, Yan S, Li B, Pan Y, Wang Y (2014) Genetic diversity and distribution of human norovirus in China (1999–2011). Biomed Res Int 13:1–15
Jothikumar N, Lowther JA, Henshilwood K, Lees DN, Hill VR, Vinje J (2005) Rapid and sensitive detection of noroviruses by using TaqMan-based one-step reverse transcription-PCR and application to naturally contaminated shellfish samples. Appl Environ Microbiol 71:1870–1875
Anderson AD, Garrett VD, Sobel J, Monroe SS, Fankhauser RL, Schwab KJ, Bresee JS, Mead PS, Higgins C, Campana J, Glass RI, Outbreak Investigation Team (2001) Multistate outbreak of Norwalk-like virus gastroenteritis associated with a common caterer. Am J Epidemiol 154(11):1013–1019
Kojima S, Kageyama T, Fukushi S, Hoshino F, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K (2002) Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods 100(1–2):107–114
Alexandros S (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34(3):772–773
Rambaut A, Lam TT, Max Carvalho L, Pybus OG (2016) Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2(1):vew007
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29(8):1969–1973
Bielejec F, Rambaut A, Suchard MA, Lemey P (2011) SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27(20):2910–2912
Lemey P, Rambaut A, Drummond AJ, Suchard MA (2009) Bayesian phylogeography finds its roots. PLoS Comput Biol 5(9):e1000520
Mori K, Motomura K, Somura Y, Kimoto K, Akiba T, Sadamasu K (2017) Comparison of genetic characteristics in the evolution of norovirus GII4 and GII17. J Med Virol 89(8):1480–1484
Galeano ME, Martinez M, Amarilla AA, Russomando G, Miagostovich MP, Parra GI, Leite JP (2013) Molecular epidemiology of norovirus strains in Paraguayan children during 2004–2005: description of a possible new GII4 cluster. J Clin Virol 58:378–384
Ayukekbong JA, Fobisong C, Tah F, Lindh M, Nkuo-Akenji T, Bergström T (2014) Pattern of circulation of norovirus GII strains during natural infection. J Clin Microbiol 52:4253–4259
Chan MCW, Hu Y, Chen H, Podkolzin AT, Zaytseva EV, Komano J, Sakon N, Poovorawan Y, Vongpunsawad S, Thanusuwannasak T, Hewitt J, Croucher D, Collins N, Vinjé J, Pang XL, Lee BE, de Graaf M, van Beek J, Vennema H, Koopmans MPG, Niendorf S, Poljsak-Prijatelj M, Steyer A, White PA, Lun JH, Mans J, Hung TN, Kwok K, Cheung K, Lee N, Chan PKS (2017) Global spread of norovirus GII.17 Kawasaki 308, 2014–2016. Emerg Infect Dis 23(8):1354–1359
de Graaf M, van Beek J, Vennema H, Podkolzin AT, Hewitt J, Bucardo F, Templeton K, Mans J, Nordgren J, Reuter G, Lynch M, Rasmussen LD, Iritani N, Chan MC, Martella V, Ambert-Balay K, Vinjé J, White PA, Koopmans MP (2015) Emergence of a novel GII.17 norovirus—end of the GII4 era? Euro Surveill 20(26):21178
Chen C, Yan JB, Wang HL, Li P, Li KF, Wu B, Zhang H (2018) Molecular epidemiology and spatiotemporal dynamics of norovirus associated with sporadic acute gastroenteritis during 2013–2017, Zhoushan Islands, China. PLoS ONE 13(7):e0200911
Han J, Wu X, Chen L, Fu Y, Xu D, Zhang P, Ji L (2018) Emergence of norovirus GIIP.16-GII.2 strains in patients with acute gastroenteritis in Huzhou, China, 2016–2017. BMC Infect Dis 18(1):342
Ji L, Chen L, Xu D, Wu X, Han J (2018) Nearly complete genome sequence of one GII17 norovirus identified by direct sequencing from HuZhou, China. Mol Genet Genomic Med 6(5):796–804
Chan MC, Lee N, Hung TN, Kwok K, Cheung K, Tin EK, Lai RW, Nelson EA, Leung TF, Chan PK (2015) Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat Commun 6:10061
Tan M, Jiang X (2014) Histo-blood group antigens: a common niche for norovirus and rotavirus. Expert Rev Mol Med 16:e5
Lindesmith LC, Debbink K, Swanstrom J, Vinjé J, Costantini V, Baric RS, Donaldson EF (2012) Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002. J Virol 86:873–883
Zhang XF, Huang Q, Long Y, Jiang X, Zhang T, Tan M, Zhang QL, Huang ZY, Li YH, Ding YQ, Hu GF, Tang S, Dai YC (2015) An outbreak caused by GII.17 norovirus with a wide spectrum of HBGA-associated susceptibility. Sci Rep 5:17687
Lindesmith LC, Kocher JF, Donaldson EF, Debbink K, Mallory ML, Swann EW, Brewer-Jensen PD, Baric RS (2017) Emergence of novel human norovirus GII.17 strains correlates with changes in blockade antibody epitopes. J Infect Dis 216(10):1227–1234
Jin M, Zhou YK, Xie HP, Fu JG, He YQ, Zhang S, Jing HB, Kong XY, Sun XM, Li HY, Zhang Q, Li K, Zhang YJ, Zhou DQ, Xing WJ, Liao QH, Liu N, Yu HJ, Jiang X, Tan M, Duan ZJ (2016) Characterization of the new GII.17 variant that emerged recently as the predominant strain in China. J Gen Virol 10:2620–2632
Bull RA, White PA (2011) Mechanisms of GII.4 norovirus evolution. Trends Microbiol 19(5):233–240
Sang S, Yang X (2018) Evolutionary dynamics of GII.17 norovirus. PeerJ 6:e4333
Matsushima Y, Ishikawa M, Shimizu T, Komane A, Kasuo S, Shinohara M, Nagasawa K, Kimura H, Ryo A, Okabe N, Haga K, Doan YH, Katayama K, Shimizu H (2015) Genetic analyses of GII.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan reveal a novel polymerase sequence and amino acid substitutions in the capsid region. Euro Surveill 20:21173
Qiao N, Ren H, Liu L (2017) Genomic diversity and phylogeography of norovirus in China. BMC Med Genomics 10:51
Kiulia NM, Mans J, Mwenda JM, Taylor MB (2014) Norovirus GII.17 predominates in selected surface water sources in Kenya. Food Chem Toxicol 6:221–231
Lu J, Fang L, Zheng H, Lao J, Yang F, Sun L, Xiao J, Lin J, Song T, Ni T, Raghwani J, Ke C, Faria NR, Bowden TA, Pybus OG, Li H (2017) The evolution and transmission of epidemic GII.17 noroviruses. J Infect Dis 23(2):316–319
Koo ES, Kim MS, Choi YS, Park KS, Jeong YS (2017) Occurrence of novel GII.17 and GII.21 norovirus variants in the coastal environment of South Korea in 2015. PLoS ONE 12(2):e0172237
Pu J, Kazama S, Miura T, Azraini ND, Konta Y, Ito H, Ueki Y, Cahyaningrum EE, Omura T, Watanabe T (2016) Pyrosequencing analysis of norovirus genogroup II distribution in sewage and oysters: first detection of GII.17 Kawasaki 2014 in oysters. Food Environ Virol 8(4):310–312
Flannery J, Keaveney S, Rajko-Nenow P, O'Flaherty V, Doré W (2012) Concentration of norovirus during wastewater treatment and its impact on oyster contamination. Appl Environ Microbiol 78(9):3400–3406
Yu Y, Cai H, Hu L, Lei R, Pan Y, Yan S, Wang Y (2015) Molecular epidemiology of oyster-related human noroviruses and their global genetic diversity and temporal-geographical distribution from 1983 to 2014. Appl Environ Microbiol 81(21):7615–7624
Wang A, Huang Q, Qin L, Zhong X, Li H, Chen R, Wan Z, Lin H, Liang J, Li J, Zhuang Y, Zhang Y (2018) Epidemiological characteristics of asymptomatic Norovirus infection in a population from oyster (Ostrea rivularis Gould) farms in southern China. Epidemiol Infect 146(15):1955–1964
Rasmussen LD, Schultz AC, Uhrbrand K, Jensen T, Fischer TK (2016) Molecular evidence of oysters as vehicle of norovirus GIIP.17-GII.17. Emerg Infect Dis 22:2024–2025
Gong HX (2016) Monitoring results of vibrio pathogenicity in shellfish in Zhoushan Islands. Prev Med 28(5):510–512 (in Chinese)
Acknowledgements
We are thankful to Dr. Xingguang Li for his suggestion to improve this work.
Funding
This work was supported by the Zhoushan Medical and Health Science Technology Project, Zhoushan, China [(2017G03), (2014C31073)] and special fund project of graduate student innovation in Jiangxi Province, China (YC2017-S093).
Author information
Authors and Affiliations
Contributions
CC, BW, and HZ performed the experiments and analyzed the data. CC and BW prepared the manuscript. RL and KFL edited the manuscript. HLW and JBY designed the study and provided funding. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Research involving human participants and/or animals
This article study does not involve human participants or animals.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Edited by Joachim Jakob Bugert.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11262_2020_1744_MOESM2_ESM.doc
TableS2. Genotype distribution of human norovirus (HuNoV) strains detected in Zhoushan Islands between 2013 and 2018. (DOC 46 kb)
Rights and permissions
About this article
Cite this article
Chen, C., Wu, B., Zhang, H. et al. Molecular evolution of GII.P17-GII.17 norovirus associated with sporadic acute gastroenteritis cases during 2013–2018 in Zhoushan Islands, China. Virus Genes 56, 279–287 (2020). https://doi.org/10.1007/s11262-020-01744-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-020-01744-6