Abstract
Canine parvovirus type 2a (CPV-2a) is a variant of CPV-2, which is a highly contagious pathogen causing severe gastroenteritis and death in young dogs. However, how CPV-2 participates in cell regulation and immune response remains unknown. In this study, persistently infected MDCK cells were generated through culture passage of the CPV-2a-infected cells for ten generations. Our study showed that CPV-2a induces cell proliferation arrest and cell morphology alternation before the fourth generation, whereas, the cell morphology returns to normal after five times of passages. PCR detection of viral VP2 gene demonstrated that CPV-2a proliferate with cell passage. An immunofluorescence assay revealed that CPV-2a particles were mainly located in the cell nuclei of MDCK cell. Then transcriptome microarray revealed that gene expression pattern of MDCK with CPV-2a persistent infection is distinct compared with normal cells. Gene ontology annotation and Kyoto Encyclopedia of Genes and Genome pathway analysis demonstrated that CPV-2a infection induces a series of membrane-associated genes expression, including many MHC protein or MHC-related complexes. These genes are closely related to signaling pathways of virus–host interaction, including antigen processing and presentation pathway, intestinal immune network, graft-versus-host disease, and RIG-I-like helicases signaling pathway. In contrast, the suppressed genes mediated by CPV-2a showed low enrichment in any category, and were only involved in pathways linking to synthesis and metabolism of amino acids, which was confirmed by qPCR analysis. Our studies indicated that CPV-2a is a natural immune activator and has the capacity to activate host immune responses, which could be used for the development of antiviral strategy and biomaterial for medicine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Canine parvovirus type 2 (CPV-2), a type species of feline parvovirus subgroup of parvovirus genus within the Parvoviridae family that emerged at the end of the 1970s as a causative agent of severe, fatal hemorrhagic gastroenteritis and death in domestic dogs [1, 2]. To date, the original CPV-2 has been completely substituted by three major genetic and antigenic variants, termed as CPV-2a, CPV-2b, and CPV-2c, identified in 1980, 1984, and 2000, respectively [3–6]. The virus has a high genomic substitution rate similar to those of RNA viruses [7] and undergoes continuous evolution [8]. Nowadays, CPV-2a is a prevalent CPV-2 field strain circulating in China [9].
CPV-2a belongs to small, nonenveloped icosahedral virus, has a single-stranded DNA genome of approximate 5.2 kb in length, which contains two major open reading frames (ORFs) [10]. The first ORF encodes two nonstructural proteins (NS1 and NS2), which are critical for viral replication and DNA packaging [10]. The second ORF encodes viral capsid proteins (VP1 and VP2), which are the main antigens responsible for inducing protective antibodies [11, 12]. VP2 is the main component to determine host range and mediate virus–receptor interaction during CPV-2 infection [13, 14].
Parvovirus infection causes different effects on host cells, which depend on the species of virus and the cell lines [15]. CPV-2 is involved in induction of apoptosis in several cell lines, including Madin-Darby canine kidney (MDCK) cells [16, 17]. However, the evidence is ambiguous and very limited. To further investigate CPV-2 infection-induced cell effect and the underlying mechanism of CPV-mediated cellular regulation, we established a persistently infected MDCK cell line with CPV-2a, isolated in China, and performed the transcriptome microarray analyses in an attempt to obtain a clearer understanding of how CPV-2a infects MDCK cells and how CPV-2a regulates host gene expression in its susceptible cells, hopefully stimulating insight into the development of antiviral strategy and identification of nanomaterial for medicine.
Materials and methods
Cells and virus
MDCK cells, obtained from China Center for Type Culture Collection (CCTCC, Wuhan, China), were maintained in the Dulbecco's modified Eagle medium (DMEM, Gibco/Life Technologies, Carlsbad, CA, USA) supplemented with 10 % (v/v) fetal bovine serum (FBS, Gibco), and grown at 37 °C with 5 % CO2. CPV-2 strain (CPV-LZ1) was isolated in China as described previously [18, 19]. The virus strain belongs to genotype CPV-2a [19]. GenBank: JQ268283.1. Virus purification and titration were monitored by Xu et al. [18].
Generation of persistent infected MDCK cell line
Monolayer MDCK cells with trypsin digestion were plated into new cell culture flasks (Nunclon, Roskilde, Denmark), supplemented with 1/10 (v/v) CPV-2a virus stock (Multiplicity of Infection, MOI = 1) in DMEM containing 5 % FBS. The infected MDCK cells were cultured at 37 °C with 5 % CO2. Cell growth and morphology were monitored daily. The cells, propagated with CPV-2a, were cultivated for ten passages. The supernatant and cells of each passage were collected and stored at −70 °C until use. The uninfected MDCK cells act as mock control.
PCR assay for CPV-2a gene detection
DNA was prepared from the supernatants by boiling the culture samples for 10 min and chilling on ice for 10 min [20]. The CPV-LZ1 solution act as positive control and uninfected MDCK cells culture act as negative control. CPV capsid protein VP2 partial segment (374 bp) was detected using semiPCR assay as previous described [21]. Meanwhile, cellular total RNA were extracted using Trizol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacture’s protocol to detect Gapdh expression level, which was used as loading control. PCR primers sequences are listed below
-
CPV-F: 5′-ATTTCTACGGGTGCTTTC-3′,
-
CPV-R: 5′-ACTTTAGTTGGTGGCTGA-3′;
-
Gapdh-F: 5′- ACGGCACAGTCAAGGCTGAG-3′,
-
Gapdh-R: 5′-CAGCATCACCCCATTTGATGTTGG-3′.
PCR was performed using a Bio-Rad Thermal Cycler (Bio-Rad Laboratories Srl, Milan, Italy) with a 25 μL reaction containing 12.5 μL of premix Taq (Takara, Dalian, China), 1 μL of primers CPV-F and CPV-R (10 μM) and 1 μL DNA samples. The following thermal system was used: 95 °C 5 min for activation of Taq DNA polymerase, then 30 cycles of 95 °C 30 s for DNA denaturation, 55 °C 30 s for primer annealing, and 72 °C 30 s for extension. The band intensity analysis was evaluated with ImageJ.
Immunofluorescence assay (IFA)
Monoclonal antibody against CPV-2a was used in IFA to evaluate the virus proliferation and cellular localization. The fourth and tenth passage of MDCK cells (MDCK F4 and F10) infected with CPV-LZ1 were harvest and fixed in 4 % paraformaldehyde for 15 min at room temperature. Then samples were treated with 0.5 % Triton X-100 for 30 min, washed with phosphate buffer saline (PBS) for three times. The fixed cells were blocked for 1 h in PBS containing 5 % new born calf serum (NBS, Hyclone, Logan, UT, USA), and incubated with an in-house mouse antiCPV-2a monoclonal antibody (diluted 1:100 in PBST with 3 % NBS) overnight at 4 °C. After washing three times in PBST, the cells were stained with TRITC-conjugated goat antimouse IgG (Sigma-Aldrich, St Louis, MO, USA, diluted 1:500 in PBST with 3 % NBS) at 37 °C for 1 h. Nuclei were stained with DAPI (Roche, Lewes, UK). Following three more washes, the plates were examined under a fluorescence microscope (Leica Microsystems, Wetzlar, German).
RNA preparation and microarray analysis
Total RNA was isolated from the tenth generation of MDCK cells with CPV-LZ1 infection using TRIzol reagent (Life Technologies) following the manufacture’s instruction. The uninfected F10 MDCK cells act as mock control. The RNA quality was checked using a NanoVue UV spectrophotometer (Thermo Scientific, Rockford, IL, USA). Qualified total RNA was further purified by RNA LabChip kit (Agilent Technologies, Santa Clara, CA, USA).
Samples RNA was amplified using a GeneChip 3‘IVT Express Kit (Affymetrix, Cleveland, OH, USA) to generate first strand cDNA and second strand cDNA, respectively. Then complementary RNA (cRNA) was produced and labeled using GeneChip HT IVT Labeling Kit (Affymetrix). The labeled cRNA was purified and quality test by an Agilent RNA 6000 Nano Kit (Agilent Technologies), followed by processed for microarray analysis on an Affymetrix Canine Genome 2.0 Array (Affymetrix) using a GeneChip Hybridization, Wash, and Stain Kit (Affymetrix), according to the manufacturer’s instructions. Arrays were scanned by an Affymetrix GeneChip Scanner 3000 System (Affymetrix). The raw data were normalized by Range Migration Algorithm and further analyzed by Affymetrix Microarray Suite Algorithm (MAS5).
Microarray data deposition
Transcriptome profiling data were submitted to the gene expression omnibus (GEO) archive following the MIAME (Minimum Information about a Microarray Experiment) instructions. The GEO accession number is GSE72397.
Real-time quantitative PCR (qPCR) analysis
For mRNA quantitative analyses, cellular total RNA was extracted using TRIzol reagent (Life Technologies) and the RNA amount was evaluated by an Epoch Multi-Volume Spectrophotometer System (BioTek, Winooski, VT, USA). For mRNA quantitative analysis, total RNA was reversed transcribed with a PrimeScript RT reagent Kit (Takara), and then qPCR was performed by the use of SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara) on an Agilent Mx3000P qPCR system (Agilent Technologies) following the manufacturer’s instructions. The comparative Ct method was used for the quantification of the target gene mRNA. The level of β-actin mRNA was used as an internal control. The qPCR conditions were shown as follow: 95 °C for 2 min, and 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Primer sequences are shown in Table 1. All primers were synthesized by Genewiz Corporation (Beijing, China).
For gene copies numbers of CPV-2a analyses, viral DNA was harvest from the supernatants of infected MDCK cells from first to tenth generation as described above. qPCR was performed on an Agilent Mx3000P qPCR system (Agilent Technologies) as described above. C t value was collected to determine viral copies.
Statistical analysis
Numerical data were collected and presented as the mean ± standard deviation (SD). Statistical analysis was performed using a two-tailed Student’s t-test. p-values of <0.05 and were considered to be significant.
Results
Evaluation of persistently infected MDCK cells
To examine the representation and characteristic of MDCK with CPV-2a infection, MDCK cells with persistent infection of CPV-2a were generated after passage for ten generations as described in Materials and Methods. Cell morphology of each passage was observed under microscope; meanwhile, cell supernatants of each generation were collected for virus VP2 gene detection by PCR assay. We found that in the early stage after CPV-2a infection, MDCK cells showed a lower cell growth rate with a stretched amorphous until the fourth generation compared to the normal MDCK (Fig. 1a, b). Another 12 h were needed for the fourth generation cells to grow to a monolayer compared to mock-infected cells (36 h). After five times of passages, MDCK proliferate and cell morphology gradually restored to normal. As to the tenth passage, MDCK showed a visible normal state under microscopy without cytopathic effect (CPE) (Fig. 1c). Cell culture supernatants of each passage were collected for CPV-2a gene detection using PCR assay. The CPV-2a viral DNA could be detected in each passage of cell supernatants, which indicated that CPV-2a proliferate as cell passage (Fig. 2a). Band intensity analysis revealed that VP2 gene level increased as cell passage until the fourth generation. Subsequently, viral gene expression gently decreased to achieve a relatively stable value till the tenth generation (Fig. 2b). In addition, we determined the gene copies numbers of CPV-2a in the supernatant of infected MDCK cells and found that CPV-2a copies increased before the fourth generation, but do not appear to add up after fourth generation, which is consistent with the result of PCR assay (Fig. 2c). Collectively, CPV-2a showed a persistent infection in MDCK cells and proliferated as cell growth, but failed to cause CPE in MDCK. After ten times of passages with cells, virus copies quantitated with cell count reached a relatively stable state in MDCK cells. These results demonstrated that steady-infected MDCK cells with CPV-2a were generated here.
CPV-2a virion detection using PCR assay. a supernatants and cell lysis of each passage of MDCK cells with CPV-2a infection were collected for DNA and mRNA preparation, respectively. CPV-2a capsid protein VP2 partial segment (374 bp) was detected using PCR assay, cellular Gapdh was used as reference. M, DNA marker; NC, supernatants of mock-treated MDCK as negative control; PC, virus stock used as positive control; 1 ~ 10, passage 1 ~ 10 of MDCK with CPV-2a infection were harvest for VP2 and Gapdh detection. b Relative intensity of each lane was determined by Image J. The column representing VP2 gene expression level were quantitated with reference to Gapdh. Data were shown as mean ± SD of triplicate experiments. Trend line was present with the histogram. c CPV-2a copies in the supernatant was showed as line chart. Data were shown as mean ± SD of triplicate experiments. dpi, days postinfection
CPV-2 replication occurs in the cell nuclei [2]. To delineate in more detail about the distribution of CPV-2a in persistent infection cells, the fourth and the tenth generation of MDCK cells with CPV-2a infection were collected to analyze CPV-2a virion localization using IFA stain. Mock-infected MDCK cells were set as control. As shown in Fig. 3, CPV-2a showed a high infection rate in MDCK, in spite of failing to cause the CPE, which also indirectly supported that MDCK is a susceptible cell line for CPV-2a. On the other side, the CPV-2a only resides in the cell nucleus, regardless of the F4 or F10 MDCK, supporting that CPV-2a replication was implemented in the cell nucleus.
CPV-2a distribution was performed using IFA assay. IFA analysis of MDCK cell with or without CPV-2a infection showed that CPV-2a particles were mainly located in cellular nuclei. CPV-2a was incubated with antiCPV-2a monoclonal antibody and stained with TRITC-conjugated second antibody (red). Nuclei were stained with DAPI (blue). a mock-treated MDCK cells. b The fourth generation of MDCK with CPV-2a infection. C, The tenth generation of MDCK with CPV-2a infection. p.i. postinfection Scale bars 50 μm (Color figure online)
Distinct gene expression signatures with CPV-2a infection
To better evaluate the feature of CPV-2a persistently infected cells and to understand the transcriptome signatures after CPV-2a infection, the tenth passage of MDCK cells with steady state infection of CPV-2a was selected for gene profiling analysis. Cellular RNA was extracted and microarray analysis was performed to identify divergent gene expression. The RNA quality monitor of RNA integrity number indicated that high-grade RNA was obtained and subsequently was used for microarray assay. Three technical replicates were performed for each sample to get reliable results. Gene profiling revealed that the CPV-2a infection caused significant divergence in gene expression. Compared with uninfected MDCK cells, under the screening criteria of a >1.5-fold change (FC) and p < 0.05, about 660 protein-coding genes showed significantly differential expression in stably infected MDCK cell, among which ~360 genes (0.83 % of total genes on the arrays) were up-regulated and ~300 genes (0.8 % of total genes on the arrays) were down-regulated (Fig. 4). The profiling data indicated that CPV-2a infection significantly altered the gene expression pattern of susceptible cells.
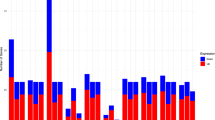
To avoid a biased interpretation of the big number of differentially expressed genes and to gain the overview of gene expression signatures, gene ontology (GO) annotation of these significantly differentially expressed genes was performed using Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) [22, 23]. We found that CPV-2a-induced genes profiling are mainly grouped in MHC proteins or MHC-related complexes (Fig. 5), which usually play roles in identifying and presenting exogenous or endogenous peptides to immune cells, and to initiate immune response [24, 25], including several central factors MHCI/II, TAP1/2, B2M, LGP2, MDA5, IRF7, and IFN-κ et al. However, the genes down-regulated by CPV-2a showed no obvious enrichment to any cellular component and biology process (Table 2). The results suggested that CPV-2a infection significantly increased a group genes expression related to antigen presentation and immune response, which may be the direct evidence that CPV-2a is another potential agent as an immune cell activator.
GO annotation of differentially expressed genes. GO enrichment analysis of the biological process (BP), cellular component (CC), and molecular function (MF) of up-regulated genes using network DAVID. Gene count of relevant BP, CC, and MF are presented as histogram, and the enrich factor, calculated by the −log p-value, are shown as line chart
CPV-2a infection activate signaling associated to immune regulation
To better understand the differentially expressed genes in the microarray data and identify specific signaling pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using DAVID after GO annotation and enrichment analysis. The results of KEGG pathway analysis revealed that these up-modulated genes, highly enriched in MHC proteins or MHC-related complexes, were involved in multiple signaling pathways associated with virus–host interaction, including antigen processing and presentation signaling, virus–host network signaling, and immune response signaling (Table 3). The most interesting signaling pathways included the well-characterized antigen processing and presentation pathway (Fig. 6) and RIG-I-like receptor (RLR) pathway (Fig. 7). Antigen processing and presentation pathway is the essential way for conversion of exogenous and endogenous proteins into immunogenic peptides via a series of proteolytic events mediated by antigen-presenting cell (APC) [26, 27], which is an important mechanism for induction of immunity to pathogen. In the CPV-2a-infected MDCK cells, many central genes of this pathway involved in MHC proteins or MHC-related complex proteins showed significant up-regulation, e.g., MHCI/II, HSP70, TAP1/2, B2 m, SLIP, and CLIP. In addition, RLR is a specific group of pattern recognition receptors (PRRs) families responsible for detecting viral pathogens and generating innate immune responses [28–30]. As shown in Fig. 7, many key regulators, such as LGP2, MDA5, IRF7, and IFN were significantly increased after CPV-2a infection, which suggested that CPV-2a could activate RLR signaling pathway.
In contrast, the down-modulated genes were only involved in pathways linking to amino acid synthesis and metabolism (Table 3), including glycine, serine, and threonine metabolism, and aminoacyl-tRNA biosynthesis, suggesting that CPV-2a infection may affect cell metabolism [31].
Taken together, these results indicated that CPV-2a infection activates multiple signaling pathways associated to host immune response, which make for activation of virus recognition and antiviral immune responses. CPV-2a infection represses some signaling related to amino acid synthesis and metabolism.
Validation of gene expression by qPCR analysis
To validate the accuracy and authenticity of our microarray data and to confirm CPV-2a-mediated signaling, we evaluated the mRNA levels of several important pathway genes with significant altered expression in the F10 cells by using mRNA qPCR. The mock-treated MDCK cells were used as control. As shown in Fig. 8, some key immune-related genes identified by the microarray were confirmed showing an obvious increasing mRNA expression level by qPCR analysis, including Major histocompatibility complex class 1 (MHC1, or DLA-12/64, a key protein involved in the presentation of foreign antigens to the immune system), Colony stimulating factor 2 (CSF2, a cytokine that stimulates the growth and differentiation of hematopoietic precursor cells from various lineages, including granulocytes, macrophages, eosinophils, and erythrocytes), Chemokine (C–C motif) ligand 2 and 28 (CCL2 and CCL28, a class of secreted cytokines involved in immunoregulatory and inflammatory processes, and Guanylate-binding protein 1 (Gbp1, an interferon-inducible protein). The down-regulated genes identified by the microarray, including Cysteinyl-tRNA synthetase (CARS), tyrosyl-tRNA synthetase (YARS), seryl-tRNA synthetase (SARS), and Regulator of G-protein signaling 2 (Rgs2) showed a remarkable decreased expression through qPCR analysis. Although the fold change was not identical, which maybe attribute to the different detection methods. These results confirmed the authenticity of microarray data and provided the further evidence that CPV-2a infection activates immune response but represses amino acid synthesis and metabolism.
Comparative analysis of qPCR test and microarray data. Relative expression levels of Gbp1, DLA-12/64, CSF2, CCL28, CCL2, SARS, CARS, YARS, and Rgs2 from microarray data and qPCR were shown as histogram and line chart. Data were represented as the mean ± SD of triplicates in three independent experiments. *p < 0.05; **p < 0.01 (Student’s t-test)
Discussion
Previous studies have shown that several parvoviruses induce cell cycle arrest [32–34]. In this study, we found that CPV-2a copies rapidly increased during early infection, cell proliferation was inhibited before the fourth generation, which indicated that CPV-2a also induces cell cycle arrest. But CPV-2a infection did not cause CPE in MDCK, just influence cell morphology gently (Fig. 1), In this study, dog-derived cell line MDCK with CPV-2a persistent infection was generated through cell passage for ten times, which can be confirmed by multiple evidences: (1) CPV-2a proliferates with MDCK cells, which was certified by PCR detection of viral VP2 gene and IFA analysis (Figs. 2, 3). (2) Cell morphology returned to normal after the fifth generation. As to the tenth passages, MDCK cells showed a visible normal state under microscopy without CPE (Fig. 1). (3) Viral amount defined by VP2 gene expression achieved relatively stable value in the tenth generation (Fig. 2). (4) CPV-2a exerts a high infection rate in MDCK via IFA stain (Fig. 3).
Notably, CPV-2a particles mainly distributed in the nucleus (Fig. 3), which was consistent with the previous reports [2], indicating that CPV-2a virion has a perfect entry mechanism into nuclei and CPV-2a replication is processed in the cell nucleus. Whereas, cytoplasm is required in accomplishment of virion assembly and release, unobservable IFA stain in this study may due to a small amount of virion are assembled and released during persistent infection.
Transcriptome profiling of MDCK cells after persistent infection of CPV-2a provide an unbiased approach to comprehensively analyze the CPV-2a-induced gene expression and regulation. The results showed that CPV-2a induces a series of differentially expressed genes. Interestingly, GO annotation showed that the up-regulated genes contains a number of membrane-associated factors, including many MHC proteins or MHC-related complexes (Fig. 5). MHC molecules are a class of membrane-associated protein, present on the surface of APCs, which identify and present exogenous or endogenous peptides to immune cells, so as to initiate immune response [24, 25]. KEGG pathway analysis revealed that these genes are closely related to signaling pathways of virus–host interaction, including antigen processing and presentation pathway, intestinal immune network, graft-versus-host disease and RLR signaling pathway (Table 3). RLRs, together with Toll-like receptors and nucleotide oligomerization domain (NOD)-like receptors, are three major receptor systems of PRRs in innate immunity for recognition of microorganisms in various cells [28] and are responsible for activation of antiviral immune responses (Fig. 6 , 7). Our qPCR data also confirmed that CPV-2a infection activates immune-related genes (Fig. 8). All these results demonstrated that CPV-2a is a great immune activation agent. Actually, many other parvoviruses infection has been reported to activate immune response in virus-induced cell lines [35–38]. Our results provided direct evidences that CPV-2a is also a potential immune cell activator. In contrast, down-modulated genes by CPV-2a are mainly involved in pathways linking to amino acid synthesis and metabolism (Table 3), including glycine, serine and threonine metabolism, and aminoacyl-tRNA biosynthesis, indicating that CPV-2a infection may affect cell metabolism.
Previous reports have demonstrated that parvovirus showed preferential replication in tumor-derived and oncogene-transformed cells, as well as onco-suppressive activity in experimental animals [35]. By the reasons of these properties, parvovirus was widely used as delivery vectors for inducing host immune responses and antitumor effects. Parvovirus-based antitumor nanotools were rapidly developed in past two decades [36–39]. As a member of parvovirus, CPV-2a causes severe disease in dogs, whereas it is not susceptible to many other species including human. Interestingly, infectious CPV particles could be confirmed to bind and enter human cells, but viral replication could not be accomplished [14]. In addition, CPV-2a has a specific cellular receptor, transferrin receptor, which is a characteristic high expressed carrier protein in tumor cells [40]. It provides the possibility to deliver drugs specifically to tumor cells utilizing CPV vector. Indeed, CPV, and CPV virus-like particles have been employed in drugs delivery and CPV is being investigated as a novel nanomaterial for delivery of genes and tumor targeting [41]. Together with our studies that CPV-2a activates the immune response signaling in susceptible cells, all these results suggested that CPV-2a is natural immune cell activator and antitumor vector. Nevertheless, several questions need further investigations. For example, which components are responsible for activation of immune-related signaling, viral structural proteins, nonstructural proteins, or nucleic acid? And how the CPV-2a activates immune signaling?
In conclusion, our studies established a MDCK cell model with CPV-2a persistent infection, and found that CPV-2a virion localized in nuclei but failed to cause CPE in MDCK cells. CPV-2a infection significantly activates host immune response-related signaling and CPV-2a is another potential immune cell activator, although further research is needed to investigate the effect on other cells lines and the underlying mechanisms. Yet, some studies mentioned that CPV-2a induces apoptosis [15, 17], but our gene profiling data showed no connection between CPV-2a infection and apoptosis, which may be attribute to the virus types. This work extends our knowledge of CPV-2a-mediated cell infection and regulation. These findings will pave ways for developing novel antiviral strategy and nanomaterial for medicine.
References
M. Battilani, S. Ciulli, E. Tisato, S. Prosperi, Virus Res. 83, 149–157 (2002)
N. Decaro, C. Buonavoglia, Vet. Microbiol. 155, 1–12 (2012)
C. Buonavoglia, V. Martella, A. Pratelli, M. Tempesta, A. Cavalli, D. Buonavoglia, G. Bozzo, G. Elia, N. Decaro, L. Carmichael, J. Gen. Virol. 82, 3021–3025 (2001)
V. Martella, N. Decaro, G. Elia, C. Buonavoglia, J. Vet. Med. B 52, 312–315 (2005)
C.R. Parrish, P.H. O’Connell, J.F. Evermann, L.E. Carmichael, Science 230, 1046–1048 (1985)
C.R. Parrish, Virology 183, 195–205 (1991)
L.A. Shackelton, C.R. Parrish, U. Truyen, E.C. Holmes, Proc. Natl. Acad. Sci. U S A 102, 379–384 (2005)
E.T. Babalola, O.K. Ijaopo, I.O. Okonko, J. Immunoassay Immunochem. 37, 16–28 (2016)
L. Yi, M. Tong, Y. Cheng, W. Song, S. Cheng, Transbound. Emerg. Dis. 63, e262 (2014)
D. Wang, W. Yuan, I. Davis, C.R. Parrish, Virology 240, 273–281 (1998)
E. Ayuso, R. Pérez, L. Calleros, A. Marandino, N. Sarute, G. Iraola, S. Grecco, H. Blanc, M. Vignuzzi, O. Isakov, N. Shomron, L. Carrau, M. Hernández, L. Francia, K. Sosa, G. Tomás, Y. Panzera, PLoS One 9, e111779 (2014)
M.S. Chapman, M.G. Rossmann, Virology 194, 491–508 (1993)
K. Hueffer, L. Govindasamy, M. Agbandje-McKenna, C.R. Parrish, J. Virol. 77, 10099–10105 (2003)
J.S. Parker, W.J. Murphy, D. Wang, S.J. O’Brien, C.R. Parrish, J. Virol. 75, 3896–3902 (2001)
J. Nykky, J.E. Tuusa, S. Kirjavainen, M. Vuento, L. Gilbert, Int. J. Nanomed. 5, 417–428 (2010)
B. Bauder, A. Suchy, C. Gabler, H. Weissenbock, J. Vet. Med. B 47, 775–784 (2000)
J. Doley, L.V. Singh, G.R. Kumar, A.P. Sahoo, L. Saxena, U. Chaturvedi, S. Saxena, R. Kumar, P.K. Singh, R.S. Rajmani, L. Santra, S.K. Palia, S. Tiwari, D.R. Harish, A. Kumar, G.S. Desai, S. Gupta, S.K. Gupta, A.K. Tiwari, Appl. Biochem. Biotechnol. 172, 497–508 (2014)
J. Xu, H.C. Guo, Y.Q. Wei, H. Dong, S.C. Han, D. Ao, D.H. Sun, H.M. Wang, S.Z. Cao, S.Q. Sun, Appl. Microbiol. Biotechnol. 98, 3529–3538 (2014)
J. Xu, H.C. Guo, Y.Q. Wei, L. Shu, J. Wang, J.S. Li, S.Z. Cao, S.Q. Sun, Transbound. Emerg. Dis. 62, 91–95 (2015)
N. Decaro, G. Elia, C. Desario, S. Roperto, V. Martella, M. Campolo, A. Lorusso, A. Cavalli, C. Buonavoglia, J. Virol. Methods 136, 65–70 (2006)
K. Rypul, R. Chmielewski, E. Smielewska-Los, S. Klimentowski, J. Vet. Med. B 49, 142–145 (2002)
W. Huang da, B.T. Sherman, R.A. Lempicki, Nat. Protoc. 4, 44–57 (2009)
W. Huang da, B.T. Sherman, R.A. Lempicki, Nucleic Acids Res. 37, 1–13 (2009)
C. Watts, Nat. Immunol. 5, 685–692 (2004)
A.L. Ackerman, P. Cresswell, Nat. Immunol. 5, 678–684 (2004)
E.S. Trombetta, I. Mellman, Annu. Rev. Immunol. 23, 975–1028 (2005)
W.R. Heath, F.R. Carbone, Nat. Rev. Immunol. 1, 126–134 (2001)
O. Takeuchi, S. Akira, Curr. Opin. Immunol. 20, 17–22 (2008)
C.B. Moore, J.P. Ting, Immunity 28, 735–739 (2008)
A. Rajput, A. Kovalenko, K. Bogdanov, S.-H. Yang, T.-B. Kang, J.-C. Kim, J. Du, D. Wallach, Immunity 34, 340–351 (2011)
X. Chen, K.H. Jhee, W.D. Kruger, J. Biol. Chem. 279, 52082–52086 (2004)
M.B. Oleksiewicz, S. Alexandersen, J. Virol. 71, 1386–1396 (1997)
A. Op De Beeck, J. Sobczak-Thepot, H. Sirma, F. Bourgain, C. Brechot, P. Caillet-Fauquet, J. Virol. 75, 11071–11078 (2001)
E. Morita, A. Nakashima, H. Asao, H. Sato, K. Sugamura, J. Virol. 77, 2915–2921 (2003)
M. Enderlin, E.V. Kleinmann, S. Struyf, C. Buracchi, A. Vecchi, R. Kinscherf, F. Kiessling, S. Paschek, S. Sozzani, J. Rommelaere, J.J. Cornelis, J. Van Damme, C. Dinsart, Cancer Gene. Ther. 16, 149–160 (2009)
K. Wetzel, S. Struyf, J. Van Damme, T. Kayser, A. Vecchi, S. Sozzani, J. Rommelaere, J.J. Cornelis, C. Dinsart, Int. J. Cancer 120, 1364–1371 (2007)
S.J. Russell, A. Brandenburger, C.L. Flemming, M.K. Collins, J. Rommelaere, J. Virol. 66, 2821–2828 (1992)
M. Malerba, L. Daeffler, J. Rommelaere, R.D. Iggo, J. Virol. 77, 6683–6691 (2003)
S. Ponnazhagan, K.A. Weigel, S.P. Raikwar, P. Mukherjee, M.C. Yoder, A. Srivastava, J. Virol. 72, 5224–5230 (1998)
J. Wen, S. Pan, S. Liang, Z. Zhong, Y. He, H. Lin, W. Li, L. Wang, X. Li, F. Zhong, Biomed. Res. Int. 2013, 172479 (2013)
P. Singh, G. Destito, A. Schneemann, M. Manchester, J. Nanobiotechnol. 4, 2 (2006)
Funding
This work was supported by grants from International Science & Technology Cooperation Program of China (2014DFA31890), the National Science and Technology Support Program (2013BAD12B00), and the Fundamental Research Funds for the Chinese Academy of Agricultural Sciences (2015ZL062).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declare no potential conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Edited by Zhen F. Fu.
Rights and permissions
About this article
Cite this article
Fan, XX., Gao, Y., Shu, L. et al. Transcriptome profiling indicating canine parvovirus type 2a as a potential immune activator. Virus Genes 52, 768–779 (2016). https://doi.org/10.1007/s11262-016-1363-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-016-1363-5