Abstract
The objective of the present study was to evaluate the epidemiology of influenza A(H1N1)pdm09 and its hemagglutinin (HA) molecular and phylogenetic analysis during 2010–2014 in Dalian, North China. A total of 3717 influenza-like illness (ILI) cases were tested by real-time PCR and 493 were found to be positive. Out of these 493 cases, 121 were subtype influenza A(H1N1)pdm09, of which 14 cases were reported in 2010–2011, 29 in 2012–2013, and 78 in 2013–2014. HA coding regions of 45 isolates were compared to that of the vaccine strain A/California/7/09(H1N1), and a number of variations were detected. P83S, S185T, S203T, R223Q, and I321V mutations were observed in all of the Dalian isolates. Furthermore, a high proportion >71 % of the strains possessed the variation D97N and K283E. Phylogenetic analysis confirmed the close match of the majority of circulating strains with the vaccine strains. However, it also reveals a trend of strains to accumulate amino acid variations and form new phylogenetic groups.
Similar content being viewed by others
Introduction
Influenza A(H1N1)pdm09 virus characterized by gene segments from avian, swine, and human origin causes the first influenza pandemic of the 21st century. It emerged in Mexico on April 2009 and spread globally within 3 months resulting in more than 18,500 deaths reported within a year [1]. The Food and Drug Administration (FDA) and the World Health Organization (WHO) recommended A/California/07/2009(H1N1) for use as the strain for the vaccines against A(H1N1)pdm09 virus [2], while, even under immune pressure, the highly active A(H1N1)pdm09 variants emerged again in December 2010 [3].
Hemagglutinin (HA) is a surface glycoproteins of influenza virus, which play an essential role in the early stages of viral replication. It binds to cellular oligosaccharide receptors containing sialic acid and mediates entry of the viral genome into the host cell by causing the fusion of host endosomal membrane with the viral envelope [4]. HA monomers are synthesized as precursors that are then glycosylated and cleaved into two smaller polypeptides: the HA1 and HA2 subunits. The continuous process of genetic change among influenza strains, which mainly occurs in the HA1, causes the emergence of new antigenic variants [5]. Since the beginning of the 2009 pandemic, A(H1N1)pdm09 continuously evolved, phylogenetic analysis of the A(H1N1)pdm09 HA gene showed that it can be clustered into eight major genetic groups [6]. Acquiring new amino acid changes may alter viral features such as antigenic characteristics, virulence, and its antiviral drug susceptibility. Thus, constant surveillance should be strengthened to monitor the pathogenicity of circulating strains.
The main objective of the present study is to evaluate the epidemiology and phylogenetic analysis of A(H1N1)pdm09 that circulated in Dalian during July 2010 to June 2014. Monitoring of antigenic sequence variations among vaccine and circulating strains is helpful to predict the effects of vaccination against the current strains and explain the rapid spread and predominance of the variants.
Materials and methods
Design
3717 nasopharyngeal swab samples from ILI (influenza-like illness) patients were collected and sent to Dalian Center for Disease Control and Prevention for pandemic influenza diagnosis between July 2010 and June 2014. The following is framework of surveillance protocol: Real-time PCR was used to detect influenza viruses RNA and their subtypes (seasonal A(H1N1), seasonal A(H3N2), and influenza A(H1N1)pdm09). Positive samples were cultured in MDCK cells and HA1 regions were amplified with RT-PCR. PCR products were purified and sequenced to compare with the HA1 regions of vaccine strain.
Detection of influenza virus types A and B and type A subtypes
Total RNA was isolated from clinical samples using RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. One-Step PrimeScript RT-PCR Kit (TaKaRa, Dalian, China) was chosen for initial detection of influenza A and B viruses and the subsequent subtyping of influenza A-positive samples. The specific primers are shown in Table 1. Multiplex real-time PCR was performed to detect types A and B and subtypes H3N2 and A(H1N1)pdm09 virus. Briefly, total reaction volume was 20 μl that contained 10 μl of One-Step RT-PCR Buffer III (2×), 0.4 μl of TaKaRa Ex TaqHS, 0.4 μl of PrimeScript RT Enzyme Mix II, 0.4 μl of ROX Reference Dye II, 0.2 mM of influenza A and B virus (or H3N2 and H1N1pdm09 virus) primers, 0.1 mM of influenza A and B virus (or H3N2 and H1N1pdm09 virus) probes, and 2 μl of RNA sample. Real-time PCR was performed to detect H1N1 virus. Briefly, total reaction volume was 20 μl, containing 10 μl of One-Step RT-PCR Buffer III (2×), 0.4 μl of TaKaRa Ex TaqHS, 0.4 μl of PrimeScript RT Enzyme Mix II, 0.4 μl of ROX Reference Dye II, 0.4 mM of H1N1 primers, 0.2 mM of H1N1 probes, and 2 μl of RNA sample. The cycling protocol was 45 °C for 5 min and 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 34 s, in an ABI 7500 Real-Time PCR System (Applied Biosystems, USA).
MDCK cell culture
The preparation of influenza strains has been described previously [7]. Briefly, flasks (T-25) with the grown Madine Darby Canine Kidney (MDCK) cells were prepared by washing two times with PBS. 500 μl of the influenza-positive specimens was inoculated onto MDCK cell culture at 35 °C under 5 % CO2 for 60 min. After removing the supernatant with a sterile pipette, 5 ml of serum-free Dulbecco’s Modified Eagles Medium (DMEM) supplemented with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (2 μg/ml) was added. The flask was incubated at 35 °C under 5 % CO2. The flasks were monitored daily for virus growth by cytopathic effects (CPE), and the cell culture supernatant was harvested when at least 75 % of the cell monolayer is exhibiting CPE. The presence of virus was assessed by a hemagglutination (HA) procedure using a 0.5 % suspension of human red blood cells group O and Rhesus negative. All cell culture reagents were obtained from Gibco BRL.
One-step RT-PCR for the amplification of HA1 genes
Total RNA was isolated from A(H1N1)pdm09-positive cell culture supernatants using RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Reverse transcription and HA1 amplification were achieved using the SuperScript™ III One-Step RT-PCR system (Invitrogen, Carlsbad, CA) with SwHA-F1 and SwHA-R primer pair (Table 1). In brief, a reaction volume of 50 μl contained 25 μl buffer (2×), 2 μl of each primer (10 μM), 1 μl of Superscript III/Platinum enzyme mix, 0.5 μl of RNase, and 5 μl of purified RNA. Cycling conditions in a Veriti 96-well thermal cycler (Applied Biosystems, Weiterstadt, Germany) were 50 °C for 30 min and 94 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 2 min. Results of 2 % agarose gel electrophoresis revealed a 1264-bp PCR product.
PCR product purification and sequencing
PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Purified PCR products were sequenced (Shenzhen Huada Gene Research Institute, China) using SwHA-F1 and SwHA-R primer pair (Table 1).
Phylogenetic analysis
45 representative sequences were selected by randomly choosing from 64 A(H1N1)pdm09 isolates during July 2010 to June 2014 of Dalian. Reference sequences of A(H1N1)pdm09 (including 1 vaccine sequence and 9 reference sequences of groups) were retrieved from Global Initiative on Sharing Avian Influenza Data (GISAID). The sequence data were analyzed with DNASTAR, Lasergene v7.1. After alignment and trimming of the sequences, the length of HA1 genes of A(H1N1)pdm09 was 981 bp. Phylogenetic tree was constructed with the neighbor-joining method, bootstrapped with 1000 replicates using MEGA software version 5.05. The GenBank accession numbers for the sequences of HA1 genes of A(H1N1)pdm09 determined in this study are KU710443-KU710487.
Results
Surveillance and prevalence of influenza virus among ILI patients
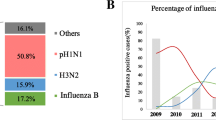
A total of 3717 influenza-like illness (ILI) cases identified at two referral hospitals were tested for influenza virus in the 4-year period, 2010–2014. Real-time PCR analysis revealed that 493 (13.3 %) of the 3717 samples were positive for influenza viruses. Out of these 493, 121 (3.3 %) were confirmed as A(H1N1)pdm09, 234 (6.3 %) were H3N2, and 138 (3.7 %) were influenza B, while none were detected as H1N1 (Table 2).
The monthly distribution of influenza virus from confirmed cases tested by real-time PCR was also studied (Fig. 1). Influenza A viruses, including H3N2, A(H1N1)pdm09, and influenza B viruses, peaked during different periods. H3N2 virus activity peaked during August–October 2010 and remained low until peaking again during January–February 2013. Influenza B virus activity was low during 2010 and peaked during January–March 2012, with two periods (January–March 2012 and January–March 2014) showing highest activity. H3N2 and influenza B persisted throughout the study period, whereas for A(H1N1)pdm09, it was emerged in 2010–2011 period, with disappearance in 2011–2012, and it is interesting to note that A(H1N1)pdm09 re-emerged and co-circulated with H3N2 during 2012–2013, becoming the dominant subtype during 2013–2014.
Phylogenetic and molecular characterization of A(H1N1)pdm09 strains
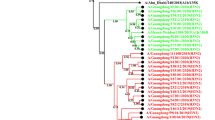
The HA1 sequences (Asp-1 to Arg-327) from 45 Dalian isolates were compared with vaccine strain A/California/7/09(H1N1) during 2010–2014. Molecular analysis shows that all viruses displayed the amino acid mutations P83S, S185ET, S203T, R223Q, and I321V in the HA1 region. Furthermore, a high proportion, 75.6 %, of the Dalian strains possessed the variation D97N. The third most common variation was K283E, which was observed at 71.1 % of the Dalian strains. Phylogenetic analysis showed Dalian circulating strains belonged to two phylogenetic groups, during 2010–2014 (Fig. 2). In detail, 34 of the 45 sequences (75.6 %) belonged to genetic group 6, characterized by the double mutation D97N and S185T. Three sub-clusters were seen within this group, 3 sequences (6.7 %) belonged to genetic group 6A, 20 sequences (44.4 %) belonged to genetic group 6B, and 11 sequences (24.4 %) belonged to genetic group 6C. Group 7, characterized by the mutations S185T, S143G, and A197T, included 11 sequences (24.4 %). In this group, eight also had the S84G and H138Q mutations. Genetic groups were named according to the European Centre for Disease Prevention and Control’s (ECDC) technical document [8–10].
Phylogenetic analysis of HA1 genes of influenza A(H1N1)pdm09 virus isolated in Dalian during July 2010–June 2014. Red boxes indicate common amino acid changes, compared to the vaccine strain, A/California/7/09. The tree was constructed with the neighbor-joining method, bootstrapped with 1000 replicates using MEGA software version 5.05 (Color figure online)
Discussion
Influenza surveillance from Dalian CDC indicates that influenza contributes significantly to the number of ILI patients seeking care in Dalian during 2010 to 2014. Overall, influenza A and B viruses predominated during distinct periods with little overlap (Fig. 1). Influenza A(H1N1)pdm09 virus spread fast and becomes a worldwide pandemic these years. In this study period, A(H1N1)pdm09 emerged with low rate during 2010–2011, disappeared during 2011–2012, re-emerged during 2012–2013 and became the dominant subtype during 2013–2014 (Table 2). The low incidence during 2010–2011 and disappearance of A(H1N1)pdm09 in 2012 may partly be explained by immune pressure, while it is becoming increasingly clear that current vaccines can only provide moderate protection against influenza and such protection is greatly reduced or absent in some seasons [11]. Some observational studies showed that median monovalent A(H1N1)pdm09 vaccine effectiveness was 69 % (range 60–93) [12, 13]. Mutations accumulated in HA genes may indirectly influence antigenicity and cause epidemic.
Most interestingly, normal seasonal rhythm of influenza epidemics was disturbed by a significant prevalence peak of H3N2 in summer 2010 of Dalian (Fig. 1). This result is broadly consistent with WHO’s 2010–2011 seasonal influenza reviews of southern China and Mexico, which showed that H3N2 was the predominant virus circulating during the northern hemisphere summer, peaking in late August to early September [14]. Dalian, a famous seaside tourist city in north China, has a large numbers domestic tourists and foreign visitors in July or August. The migration of this scale may contribute to the initiation of influenza epidemics in summer of Dalian.
Previous pandemics showed the novel virus subtype may replace previously circulating viruses [15]. For example, in 1957, when the H2N2 strain emerged, it replaced the previously circulating H1N1 strain. Then in 1968, the H2N2 strain was replaced by the H3N2 strain [16–18]. In this study, we found that after A(H1N1)pdm09 emerged in 2009, H1N1, which was the predominant subtype in some years of Dalian, disappeared (Table 2). It looks like A(H1N1)pdm09 virus completely replaced H1N1 virus circulating. These observations are similar to situations in other parts of the world [19, 20] and correspond to the hypotheses that the emergence of novel viruses has often been coupled with the disappearance of existing seasonal virus strains [21, 22].
Gradual minor point mutations in the genes responsible for encoding the HA proteins, called antigenic drift, may occur. These point mutations may cause the emergence of new antigenic variants, which make viruses to either escape antibodies acquired by host during previous infections or to develop drug-resistant strains [23, 24]. El Moussi et al. showed that, in Tunisia, 11 out 17 (64.7 %) S185T mutation strains were from severe cases, while whether this substitution affects receptor-binding ability of the A(H1N1)pdm09 remains to be determined [25]. Mutation S203T, which located at the antigenic site Ca, has been reported to affect the infectivity and transmissibility of influenza virus in humans [26]. Melidou et al. suggested that the occurrence of 321I in severe cases could only be an effect of changing frequency over time rather than an association to severity [27]. The D222G mutation, which has been observed in severe cases previously, allows the virus to gain double specificity in receptor binding and contributes the virus to reach the lower respiratory tract [28, 29]. In this study, none of the Dalian strains showed the D222G change, while all of the viruses analyzed displayed the amino acid changes P83S, S185T, S203T, R223Q, and I321V in the HA1 region and a high proportion >71 % of the strains possessed the variation D97N and K283E. Furthermore, sporadic aa substitutions and the presence of variation in glycosylation sites within HA1 were also seen in A(H1N1)pdm09 virus. It is possible that these changes, accumulating over time, may result in a virus with different antigenic properties.
H1-based phylogenetic analysis showed that all the strains clustered in two main branches with a significant genetic distance. Approximately, 75.6 % of the strains fell into group 6 and 24.4 % into group 7. During 2010–2011, all Dalian circulating strains belonged to phylogenetic group 7. During 2012–2013, circulating strains were scattered in group 6A, 6B, 6C, and group 7, contained specific mutations in HA1 (Fig. 2). Although only one strain A/Dalian/SWL1338/2013 scattered in group 6B during 2012–2013, during 2013–2014 all the 20 Dalian strains belonged to phylogenetic group 6B. A relatively low match with the vaccine strains during 2013–2014.
The present work is the first molecular study of the influenza pandemic viruses circulating in Dalian during 2010–2014, which contribute to a better understanding of the genetic evolution of the A(H1N1)pdm09 viruses. Nevertheless, detailed genetic information of the evolution of influenza virus genes and the function of the specific mutations identified in this study is needed to clarify.
References
F.S. Dawood, A.D. Iuliano, C. Reed, M.I. Meltzer, D.K. Shay, P.Y. Cheng, D. Bandaranayake, R.F. Breiman, W.A. Brooks, P. Buchy et al., Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation. Lancet. Infect. Dis. 12(9), 687–695 (2012)
Centers for Disease Control and Prevention (CDC), Use of influenza A (H1N1) 2009 monovalent vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR. Recomm. Rep. 58, 1–8 (2009)
J. Li, T.J. Shao, X.F. Yu, J.C. Pan, X.Y. Pu, H.Q. Wang, Y. Kou, Molecular evolution of HA gene of the influenza A H1N1 pdm09 strain during the consecutive seasons 2009–2011 in Hangzhou, China: several immune-escape variants without positively selected sites. J. ClinVirol. 55(4), 363–366 (2012)
B.S. Hamilton, G.R. Whittaker, S. Daniel, Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses. 4(7), 1144–1168 (2012)
N. Sriwilaijaroen, Y. Suzuki, Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 88(6), 226–249 (2012)
I.G. Barr, C. Russell, T.G. Besselaar, N.J. Cox, R.S. Daniels, WHO recommendations for the viruses used in the 2013–2014 Northern Hemisphere influenza vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from October 2012 to January 2013. Vaccine. 32(37), 4713–4725 (2014)
K.J. Szretter, A.L. Balish, J.M. Katz, Influenza: propagation, quantification, and storage. Current protocols in microbiology, chapter 15, vol. 1 (Wiley, New Jersey, 2006), p. 15G.11.11-15G.11.22
European Centre for Disease Prevention and Control. Influenza virus characterisation, summary Europe. (2011)
European Centre for Disease Prevention and Control. Influenza virus characterisation, summary Europe, March. (2014)
European Centre for Disease Prevention and Control. Influenza virus characterisation, summary Europe, December. (2014)
M.T. Osterholm, N.S. Kelley, A. Sommer, Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 12(1), 36–44 (2012)
M.G. Thompson, L.Z. Sokolow, O. Almendares, K. Openo, M.M. Farley, J. Meek, J. Ray, P.D. Kirley, A. Reingold et al., Effectiveness of nonadjuvanted monovalent influenza A(H1N1)pdm09 vaccines for preventing reverse transcription polymerase chain reaction-confirmed pandemic influenza hospitalizations: case-control study of children and adults at 10 US influenza surveillance network sites. Clin. Infect. Dis. 57(11), 1587–1592 (2013)
P. Hardelid, D.M. Fleming, J. McMenamin, N. Andrews, C. Robertson, P. SebastianPillai, J. Ellis, W. Carman, T. Wreghitt, J.M. Watson et al., Effectiveness of pandemic and seasonal influenza vaccine in preventing pandemic influenza A(H1N1)2009 infection in England and Scotland 2009–2010. Euro. Surveill. 16(2), 19763 (2011)
World Health Organization (WHO), Summary review of the 2010–2011 northern hemisphere winter influenza season. Wkly. Epidemiol. Rec. 86, 221–232 (2011)
J.D. Mathews, J.M. Chesson, J.M. McCaw, J. McVernon, Understanding influenza transmission, immunity and pandemic threats. Influenza. Respi. Viruses. 3(4), 143–149 (2009)
C. Viboud, T. Tam, D. Fleming, A. Handel, M.A. Miller, L. Simonsen, Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine. 24(44–46), 6701–6707 (2006)
P. Palese, M.L. Shaw, Orthomyxoviridae: the viruses and their replication, in Fields virology, vol. 2, 5th edn., ed. by D.M. Knipe, P.M. Howley (Lippincott Williams & Wilkins, Philadelphia, 2007), pp. 1647–1689
R.G. Webster, W.J. Bean, O.T. Gorman, T.M. Chambers, Y. Kawaoka, Evolution and ecology of influenza A viruses. Microbiol. Rev. 56(1), 152–179 (1992)
S. Broor, A. Krishnan, D.S. Roy, S. Dhakad, S. Kaushik, M.A. Mir, Y. Singh, A. Moen, M. Chadha, A.C. Mishra et al., Dynamic patterns of circulating seasonal and pandemic A(H1N1)pdm09 influenza viruses from 2007-2010 in and around Delhi, India. PLoS. One. 7(1), e29129 (2012)
J.W. Tang, C.K. Lee, H.K. Lee, T.P. Loh, L. Chiu, P.A. Tambyah, E.S. Koay, Tracking the emergence of pandemic influenza A/H1N1/2009 and its interaction with seasonal influenza viruses in Singapore. Ann. Acad. Med. Singap. 39(4), 291–294 (2010)
P. Palese, T.T. Wang, Why do influenza virus subtypes die out? a hypothesis. MBio. 2(5), e00150 (2011)
N. Pica, R. Hai, F. Krammer, T.T. Wang, J. Maamary, D. Eggink, G.S. Tan, J.C. Krause, T. Moran, C.R. Stein et al., Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc. Natl. Acad. Sci. USA. 109(7), 2573–2578 (2012)
M.C. Zambon, Epidemiology and pathogenesis of influenza. J. Antimicrob. Chemother. 44, 3–9 (1999)
F. Carrat, A. Flahault, Influenza vaccine: the challenge of antigenic drift. Vaccine. 25(39–40), 6852–6862 (2007)
A. El Moussi, M.A. Ben Hadj Kacem, F. Pozo, J. Ledesma, M.T. Cuevas, I. Casas, A. Slim, Genetic diversity of HA1 domain of heammaglutinin gene of influenza A(H1N1)pdm09 in Tunisia. Virol. J. 10, 150 (2013)
C. Pan, B. Cheung, S. Tan, C. Li, L. Li, S. Liu, S. Jiang, Genomic signature and mutation trend analysis of pandemic (H1N1) 2009 influenza A virus. PLoS. One. 5(3), e9549 (2010)
A. Melidou, G. Gioula, M. Exindari, D. Chatzidimitriou, E. Diza, N. Malisiovas, Molecular and phylogenetic analysis of the haemagglutinin gene of pandemic influenza H1N1 2009 viruses associated with severe and fatal infections. Virus. Res. 151(2), 192–199 (2010)
J.A. Vazquez-Perez, P. Isa, D. Kobasa, C.E. Ormsby, D.P. Romero-Rodriguez, C. Ranadheera, Y. Li, N. Bastien, C. Embury-Hyatt et al., A(H1N1)pdm09HA D222 variants associated with severity and mortality in patients during a second wave in Mexico. Virol. J. 10, 41 (2013)
P.K. Chan, N. Lee, G.M. Joynt, K.W. Choi, J. Cheung, A.C. Yeung, P. Lam, R. Wong, B.W. Leung, H.Y. So et al., Clinical and virological course of infection with haemagglutinin D222G mutant strain of 2009 pandemic influenza A (H1N1) virus. J. Clin. Virol. 50(4), 320–324 (2011)
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 31201879). We sincerely thank Dr. Asraf Hussain Hashmi for his valuable comments and discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Edited by Paul Schnitzler.
Rights and permissions
About this article
Cite this article
Han, Y., Sun, N., Lv, Qy. et al. Molecular epidemiology and phylogenetic analysis of HA gene of influenza A(H1N1)pdm09 strain during 2010–2014 in Dalian, North China. Virus Genes 52, 606–612 (2016). https://doi.org/10.1007/s11262-016-1358-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-016-1358-2