Abstract
Growth traits in livestock animals are quantitative parameters, which are often controlled by many genes including growth hormone (GH) gene. However, the evidence of effect of GH gene on growth traits of cattle is poorly understood. Hence, the objective of the study was to systematically investigate the literature on single nucleotide polymorphisms (SNPs) of GH gene and their association with growth traits in cattle from four databases Google Scholar, PubMed, ScienceDirect, and Web of Science. The results indicated that fifteen (n = 15) articles with 27% of them from Indonesia qualified to be used in this study after screening. The results revealed five SNPs (1047T > C, 1180 C > T, 86,273,136 A/G, 3338 A > G and 4251 C > T) occurred across multiple investigated breeds with no common identified SNPs. Six articles observed a significant difference (p < 0.05) between growth traits and genotypes of identified SNPs. The findings showed that 7 articles (47%) investigated body weight (BW) with 6 (40%) of them found non-significant and 1 (7%) found a significant association with genotypes of the identified SNPs (3338 A > G). While 7 articles (47%) investigated weaning weight (WW) with 5 (33%) of them revealed a non-significant and 2 (13%) found a significant association with genotypes of identified SNPs (3338 A > G and 4251 C > T). This study shows that there is a lack of evidence on effect of growth hormone gene on growth traits in cattle. However, more studies are recommended for further validation of the identified SNPs and effect of growth hormone gene on growth traits in cattle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Growth Hormone (GH) is a protein hormone that is manufactured and secreted by the part of the pituitary gland (Agung et al. 2017). The hormone encoded by the GH gene and affects the productivity in beef cattle (Agung et al. 2017). Furthermore, the GH is necessary for tissue growth, fat metabolism and normal body growth (Agung et al. 2017). Agung et al. (2017) reported that the function of the GH is to play a role in reproduction traits such superovulation response, ovulation rate, fertility rate and embryo quality. The significance of the function of GH gene is to make this gene as one of candidate genes for marker assisted selection (MAS) program. Nowadays, molecular technology based on MAS has been widely used in cattle genetic improvement (Fathoni et al. 2019). Single nucleotide polymorphisms (SNPs) of GH gene and their association with growth traits in cattle has been investigated in different cattle breeds including Sumba Ongole (Agung et al. 2017), Holstein heifers (Arango et al. 2014), Colombian Creote cattle (Martinez et al. 2016), Pasundan cows (Putra et al., 2019) and Brangus bull (Thomas et al. 2007). However, based on authors’ knowledge there is no literature that has systematically investigated the SNPs of GH gene and their relationship with growth traits in cattle. To close the identified knowledge gap, the objective of the study was to systematically investigate the literature on single nucleotide polymorphisms of GH gene and their association with growth traits in cattle. This systematic review might help researchers and cattle farmers to know the genetic markers of GH gene that might be used for selection during breeding to improve growth traits in cattle.
Materials and methods
Eligibility criteria
Identification of Population, Exposure, and Outcomes (PEO) components of the research question as explained by Bettany-Saltikov (2010) was performed prior conducting the systematic review. Thereby the population was defined as “Cattle”, with an exposure of “Polymorphisms” and outcomes of “Growth Traits”. A pilot search of the PEO components on the Google Scholar database was conducted prior deciding to conduct the study.
Literature search
The literature search for the research publications was done by all the authors through the use of four database Google Scholar, PubMed, ScienceDirect, and Web of Science up to 04 December, 2023 where the following keywords were made use of: ‘growth hormone gene (GH)’, ‘single nucleotide polymorphisms/genetic effect/polymorphisms/genetic variations’, ‘growth traits’ and ‘cattle’.
Inclusion criteria
Titles and abstracts found using the search strategy were manually screened to recognize articles that were potentially appropriate. Articles were considered for inclusion in the systematic review given: some they included associations between growth hormone gene and growth traits in cattle.
Exclusion criteria
Articles published in any other language other than English, only have abstracts accessible, only have English abstract, articles talking about different species other than cattle were excluded. Lastly, duplicate articles from the different database used were also excluded.
Data extraction
The content was extracted independently by the authors. The contents extracted from the articles include the name of the 1st author, year the article was published, country, cattle breed, population size, investigated trait and genotyping method.
Ethical considerations
Ethical consideration including plagiarism, misconduct, informed consent, data falsification, and fabrication were considered by all the authors.
Results
Literature searched results
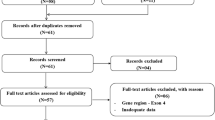
As indicated in Fig. 1, the articles retrieved for this systematic review amounted to a total of fifty-three (n = 53) from the following databases: Google Scholar (n = 28), PubMed (n = 5), Science Direct (n = 10), and Web of Science (n = 10). The duplicates (n = 19) that occurred in between the search databases were removed and the remaining articles were further analysed for inclusion and exclusion criteria. Finally, thirty four articles (n = 34) were screened on the basis of title, followed by the screening for the abstract, thereby eighteen (n = 18) articles were assessed for eligibility. A total of fifteen (n = 15) articles was used in writing this systematic review.
Characteristics of included articles
About fifteen (n = 15) included articles of the fifty-three (n = 53) analysed were retained for inclusion in the literature review (Table 1). The retained articles ranged from the year 2003 to 2020. All included articles investigated growth hormone gene and its association with growth traits in cattle. The Angus cattle breed being most investigated (Ge et al. 2003; Kayumov et al. 2019; Ruban et al. unknown) accounting for 20% (n = 3) of the included articles.
Origin of the publications
Article distribution per country is displayed in Fig. 2. All the included articles showed the origin of the publications. The results indicated that the included articles were from different countries such as Indonesia, Colombia, Mexico, Russia, United State, Thailand, Ukraine, and China. In addition, the findings revealed that the majority (27%) of the included articles were from Indonesia (Agung et al. 2017; Fathoni et al. 2019; Putra et al. 2019; Maskur & Arman 2014) followed by (13%) Colombia (Arango et al. 2014; Martinez et al. 2016), Mexico (Garrett et al. 2008; Thomas et al. 2007), Russia (Kayumov et al. 2019; Sedykh et al. 2020) and United State (Beauchemin et al. 2006; Ge et al. 2003), while the least (7%) included articles were from Thailand (China et al. 2007), Ukraine (Ruban et al. unknown), and China (Zhang et al. 2011).
Distribution of the articles by known and unknown years
Article distribution per year is presented in Fig. 3. The results revealed that fourteen articles indicated the year of publication while only one article did not reveal the year of publication. Moreover, the findings of the review indicated that included articles were published in the year 2003 (6.66%), 2006 (6.66%), 2007 (13.33%), 2008 (6.66%), 2011 (6.66%), 2014 (13.33%), 2016 (6.66%), 2017 (6.66%), 2019 (20%), 2020 (6.66%) and unknown (6.66%). The results showed that most included articles were published in year 2019 (Fathoni et al. 2019; Putra et al. 2019; Kayumov et al. 2019).
Publications by journals
Article distribution per journals is displayed in Table 2. The findings indicated that included articles were published in different journals such as Journal of the Indonesian Tropical Animal Agriculture, Genetics and Molecular Research Online Journal, Kasetsart Journal of Natural Science, Journal of Animal Science, Advances in Intelligent Systems Research, Tropical Animal Science Journal, Italian Journal of Animal Science, unknown, Iranian Journal of Applied Animal Science and Research in Veterinary Science. The results showed that most of the included articles were published by the Genetics and Molecular Research Online Journal with 27% (Arango et al. 2014; Beauchemin et al. 2006; Martinez et al. 2016; Thomas et al. 2007).
Distribution of the articles by genotyping methods
Article distributions by genotyping methods are presented in Fig. 4. The results indicated that Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) and Polymerase Chain Reaction-Single Strand Conformation Polymorphism (PCR-SSCP) were the genotyping methods used by the included articles. The results showed that most included articles 87% (n = 13) used the PCR-RFLP method for genotyping while 13% (n = 2) of the included articles (China et al. 2007; Zhang et al. 2011) used PCR-SSCP genotyping method.
Articles by genomic regions
Article distributions by genomic regions are displayed in Fig. 5. The findings indicated that only 53% (n = 8) of the included articles identified the genomic regions while the remaining 47% of the included articles did not indicate the genomic regions. Furthermore, the results showed that two (13%) (Agung et al. 2017; Arango et al. 2014) out of 15 included articles found SNPs in intron 3 and in exon 2 (Zhang et al. 2011; Martinez et al. 2016).
Identified single nucleotide polymorphisms
The identified and non-identified single nucleotide polymorphisms (SNPs) and their regions are presented in Table 3. The results indicated that out of 15 included articles, seven (47%) of them identified SNPs. Three articles identified AG SNPs (Garrett et al. 2008; Putra et al. 2019; Maskur & Arman 2014).
Genotypic frequencies
Table 3 indicates the genotypic and allelic frequencies from included articles. The findings showed that out of 15 included articles, 11 (73%) of them identified three genotypes per SNP while 4 (27%) articles (cite) did not identify genotypes per SNP. The results indicated that the genotypic frequencies from the reviewed articles ranged from 0.00 to 102.00.
Allelic frequencies
Identified allelic frequencies from different reviewed articles are presented in Table 4. Out of 15 collected reviewed articles, 13 (87%) of them were clear about allelic frequencies, whereas 2 (13%) were not. The discovered allelic frequencies ranged from the reviewed articles ranged from 0.11 to 94.10.
Association between identified SNPs’ genotypes and growth traits
The SNPs’ genotypes and their association with growth traits are detailed in Table 5. Out of a total of 15 articles reviewed, 6 articles observed a significant difference (p < 0.05) in one or more traits of interest as a result of the SNPs that occur in the GH gene of the cattle. From the 6 articles that were studied 5 traits which included average daily gain (ADG), body weight (BW), weaning weight (WW), yearling weight (YW) and body length (BL), their association with SNPs identified were investigated. About 7 articles (47%) investigated BW with 6 (40%) non-significant and 1 (7%) significantly associated with genotypes. While 7 articles (47%) investigated WW with 5 (33%) non-significant and 2 (13%) significantly associated with genotypes. Three articles (20%) investigated YW with 1 (7%) non-significant and 2 (13%) significantly associated with genotypes. Five articles (33%) investigated ADG with 3 (20%) non-significant and 2 (13%) significantly associated with genotypes. Lastly, one article (7%) investigated BL with significantly associated with genotypes.
Discussion
Genes involved in controlling the manifestation of growth traits might be interesting as candidate markers for livestock’s performance traits (Ge et al. 2003; Beauchemin et al. 2006). Growth traits are important measurable traits that have a remarkable effect on the beef cattle production industry (China et al. 2007). Understanding the effects of GH gene on growth traits of cattle is important especial when farmers want to develop management strategies focusing on genetic improvement of cattle (Zhang et al. 2011). This systematic review was performed to assess the literature on single nucleotide polymorphisms and their association with growth traits in cattle. A total fifteen (n = 15) articles of the fifty-three (n = 53) retrieved articles were retained for inclusion and used for development of this systematic review. The findings of this review showed that Angus cattle breed was the most investigated breed (Ge et al. 2003; Kayumov et al. 2019; Ruban et al. unknown) which accounting for 20% of the included articles. This might be a result that the Angus cattle breed is the most dominating popular breed found in the15 countries used on this systematic review. However, in South Africa (SA) the Angus cattle breed is not one of the common breeds reared by communal farmers. Furthermore, the used articles revealed that the 27% of the included articles were from Indonesia followed by 13% of them from Mexico, Russia and United State. This could be due to the availability of resources to perform molecular genetics studies. The results showed that most included articles (87%) used the PCR-RFLP method for genotyping while 13% of the included articles used PCR-SSCP genotyping method (China et al., 2007; Zhang et al., 2011). This could be because the PCR-RFLP genotyping method is among commonly available cheapest genotyping methods in the world (Zhang et al. 2011). Five SNPs (1047T > C, 1180 C > T, 86,273,136 A/G, 3338 A > G and 4251 C > T) were identified from the 15 included articles. The observed results indicated that out of 15 articles used in this systematic review, six (40%) articles showed a significant association between the genotypes of the observed SNPs and growth traits in cattle. Agung et al. (2017) identified 1047T > C SNP and found it to be insignificantly associated with body weight, weaning weight and yearling weight in Sumba Ongole cattle. The SNP 1180 C > T identified by Fathoni et al. (2019) in Kebumen Ongole Grade cattle, showed no significance association with studied traits. The 86,273,136 A/G SNP investigated by Garrett et al. (2008) in Brangus cattle showcased a no significant association with body weight, weaning weight and average daily gain. While Putra et al. (2019) further identified significant correlation between the 3338 A > G SNP and growth traits (weaning weight and body length). On the other hand, Zhang et al. (2011) identified significant correlation between the 4251 C > T SNP and yearling weight in Nanyang cattle. The implication of this systematic review is that the identified SNPs can be used to improve economic important growth traits in cattle breeds. The strength of this systematic review is that to the best of our knowledge there is no similar studies on systematic review of the effects of GH gene on growth traits in cattle. Hence, no studies were used for comparison of the findings. The contribution of this systematic review to the body of knowledge is to suggest that there is a relationship exists between GH gene and some of the growth traits in different cattle breeds. The limitations of this systematic review are (1) some of the articles did not mention the sex of the cattle used which makes it difficult to make the conclusion per sex and (2) no similar SNPs were identified in the included articles to proceed to meta-analysis. This systematic review suggests that there is insufficient evidence on the association between SNPs of GH gene and their association with growth traits in cattle. Therefore, there is a need to conduct more studies that will focus on investigating the relationship between SNPs of GH gene and growth traits in cattle to provide more information and validate these findings to assist on identifying genetic markers that might be used to improve growth traits of cattle during breeding programs.
Conclusion
In the current study, it was found that the growth hormone gene affects some of the growth traits of cattle and the reported SNPs might be used as possible genetic markers for selection to improve growth traits in cattle during breeding programs. Lastly, the findings of the current study shows that the identified 3338 A > G SNP may be used as a genetic marker for improving weaning weight and body length in during breeding of Pasundan cows.
Data availability
All data generated during this study are available through a request to the corresponding author.
References
Agung, P.P., Anwar, S, Putra, W.P.B., Zein, M.S A., Wulandari, A.S., Said, S. & Sudiro, A., 2017. Association of growth hormone (GH) gene polymorphism with growth and carcass in Sumba Ongole (SO) cattle. Journal of the Indonesian Tropical Animal Agriculture. 42(3):153–159, https://doi.org/10.14710/jitaa.42.3.153-159
Arango, J., Echeverri, J.J. & López, A., 2014. Association between a polymorphism in intron 3 of the bovine growth hormone gene and growth traits in Holstein heifers in Antioquia. Genetics and Molecular Research Online Journal. DOI https://doi.org/10.4238/2014.August.15.1
Bettany-Saltikov, J., (2010): Learning how to undertake a systematic review: Part 2. Nursing Standard 24, 47–56.
Beauchemin, V.R., Thomas, M.G., Franke, D.E. & Silver, G.A., 2006. Evaluation of DNA polymorphisms involving growth hormone relative to growth and carcass characteristics in Brahman steers. Genetics and Molecular Research Online Journal.
China, S., Supamit, M., Voravit, S., Panwadee, S., Skorn, K. and Sornthep T., 2007. Effect of Genetic Polymorphism of Bovine Growth Hormone Gene on Preweaning Growth Traits in a Thai Multibreed Beef Population. Kasetsart Journal of Natural Science. 41: 484–492.
Fathoni, A., Maharani, D., Aji, R. N., Choiri, R. and Sumadi, S., 2019. Polymorphism of the SNP g. 1180 C > T in leptin gene and its association with growth traits and linear body measurement in Kebumen Ongole Grade cattle. Journal of the Indonesian Tropical Animal Agriculture. 44(2):125–134. https://doi.org/10.14710/jitaa.44.2.125-134
Garrett, A. J., Rincon, G., Medrano, J. F., Elzo, M.A., Silver, G.A. & Thomas, M.G., 2008. Promoter region of the bovine growth hormone receptor gene: Single nucleotide polymorphism discovery in cattle and association bwith performance in Brangus bulls. Journal of Animal Science. 86:3315–3323. https://doi.org/10.2527/jas.2008-0990
Ge, W., Davis, M. E., Hines, H.C., Irvin, K.M. & Simmen, R.C.M., 2003. Association of single nucleotide polymorphisms in the growth hormone and growth hormone receptor genes with blood serum insulin-like growth factor I concentration and growth traits in Angus cattle. Journal of Animal Science. 1:641–648.
Kayumov, F., Kosilov, V., Gerasimov, N. & Bykova, O., 2019. The effect of SNP polymorphisms in growth hormone gene on weight and linear growth in crossbred Red Angus × Kalmyk heifers. Advances in Intelligent Systems Research.
Martinez, R., Rocha, J.F., Bejarano, D., Gomez, Y., Abuabara, Y. & Gallego, J., 2016. Identification of SNPs in growth-related genes in Colombian creole cattle. Genetics and Molecular Research Online Journal. DOI https://doi.org/10.4238/gmr.15038762
Maskur, R. & Arman, C., 2014. Association of a Novel Single Nucleotide Polymorphism in Growth Hormone Receptor Gene with Production Traits in Bali Cattle. Italian Journal of Animal Science. volume 13:3461.
Putra, W.P.B., Agung, P.P., Anwar, S. & Said, S., 2019. Polymorphism of Bovine Growth Hormone Receptor Gene (g.3338A > G) and Its Association with Body Measurements and Body Weight in Pasundan Cows. Tropical Animal Science Journal. 42(2):90–96. https://doi.org/10.5398/tasj.2019.42.2.90
Ruban, S.Y., Fedota, A. M., Lysenko, N. G., Kolisnyk, A.I. & Goraichuk, I.V., unknown. The effects of polymorphisms in calpain calpastatin and growth hormone genes on growth traits in Angus cows.
Sedykh, T.A., Dolmatova, I.Y., Valitov, F.R., Gizatullin, R.S. & Kalashnikova, L.A., 2020. The Influence of Growth Hormone Gene Polymorphism on Growth Rate of Young Cattle. Iranian Journal of Applied Animal Science. 10(3), 445–451.
Thomas, M.G., Enns, R.M., Shirley, K.L., Garcia, M.D., Garrett, A.J. & Silver, G.A., 2007. Associations of DNA polymorphisms in growth hormone and its transcriptional regulators with growth and carcass traits in two populations of Brangus bulls. Genetics and Molecular Research Online Journal. 6 (1): 222–237.
Zhang, B., Zhao, G., Lan, X., Lei, C., Zhang, C. & Chen, H., 2011. Polymorphism in GHRH gene and its association with growth traits in Chinese native cattle. Research in Veterinary Science. 92: 243–246.
Funding
The authors would like to thank the National Research Foundation (NRF) reference number (MND210415594902) for its financial support.
Open access funding provided by University of Limpopo.
Author information
Authors and Affiliations
Contributions
Initial idea of the study was by Lubabalo Bila and Thobela Louis Tyasi. All the authors contributed on the designing of the study. Lubabalo Bila wrote the first draft of the manuscript. Dikeledi Petunia Malatji and Thobela Louis Tyasi revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
It is declared by the authors that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bila, L., Malatji, D.P. & Tyasi, T.L. Single nucleotide polymorphisms of growth hormone gene in cattle and their association with growth traits: a systematic review. Trop Anim Health Prod 56, 141 (2024). https://doi.org/10.1007/s11250-024-03985-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-024-03985-1