Abstract
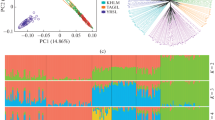
Bawri or Garri, a non-descript cattle population managed under an extensive system in Madhya Pradesh state of India, was identified and characterized both genetically and phenotypically to check whether or not it can be recognised as a breed. The cattle have white and gray colour and are medium sized with 122.5 ± 7.5 cm and 109.45 ± 0.39 cm height at withers in male and female, respectively. Double-digest restriction site associated DNA (ddRAD) sequencing was employed to identify ascertainment bias free SNPs representing the entire genome cost effectively; resulting in calling 1,156,650 high quality SNPs. Observed homozygosity was 0.76, indicating Bawri as a quite unique population. However, the inbreeding coefficient was 0.025, indicating lack of selection. SNPs found here can be used in GWAS and genetic evaluation programs. Considering the uniqueness of Bawri cattle, it can be registered as a breed for its better genetic management.


Similar content being viewed by others
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author (Mail ID: jeyvet@gmail.com) on reasonable request.
References
Catchen, J. M., Amores, A., Hohenlohe, P., Cresko, W., & Postlethwait, J. H. (2011). Stacks: building and genotyping loci de novo from short-read sequences. G3: Genes| genomes| genetics, 1(3), 171–182. https://doi.org/10.1534/g3.111.000240
Czech B, Frąszczak M, Mielczarek M, Szyda J (2018). Identification and annotation of breed-specific single nucleotide polymorphisms in Bos taurus genomes. PLoS One 13 (6). https://doi.org/10.1371/journal.pone.0198419
DAHD, 2022. Breed-wise report of livestock and poultry. Based on 20th livestock census (2022). https://dahd.nic.in/sites/default/filess/Breed%20Report%20Full%20Book.pdf (Accessed on 25-November-2023)
Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156-2158.
De Donato, M., Peters, S. O., Mitchell, S. E., Hussain, T., & Imumorin, I. G. (2013). Genotyping-by-sequencing (GBS): a novel, efficient and cost-effective genotyping method for cattle using next-generation sequencing. PloS one, 8(5), e62137.
Devadasan MJ, Kumar DR, Vineeth MR, Choudhary A, Surya T, Niranjan SK, Verma A, Sivalingam J (2020) Reduced representation approach for identification of genome-wide SNPs and their annotation for economically important traits in Indian Tharparkar cattle. 3 Biotech 10 (7):309. https://doi.org/10.1007/s13205-020-02297-z
Dickerson, G. E. (1955). Genetic slippage in response to selection for multiple objectives. In Cold Spring Harbor Symposia on Quantitative Biology (Vol. 20, pp. 213–224). Cold Spring Harbor Laboratory Press. https://doi.org/10.1101/SQB.1955.020.01.020
Dixit, S. P., Bhatia, A. K., Ganguly, I., Singh, S., Dash, S., Sharma, A., ... & Jayakumar, S. (2021). Genome analyses revealed genetic admixture and selection signatures in Bos indicus. Scientific Reports, 11(1), 1–11.
FAO. (2012). Phenotypic characterization of animal genetic resources. Animal Production and Health Guidelines No. 11. (available at www.fao.org/docrep/015/i2686e/i2686e00.pdf)
Gorlov, I. P., Gorlova, O. Y., Sunyaev, S. R., Spitz, M. R., & Amos, C. I. (2008). Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. The American Journal of Human Genetics, 82(1), 100-112.
Gurgul A, Miksza-Cybulska A, Szmatoła T, Jasielczuk I, Piestrzyńska-Kajtoch A, Fornal A, Semik-Gurgul E, Bugno-Poniewierska M (2019) Genotyping-by-sequencing performance in selected livestock species. Genomics 111 (2):186-195. https://doi.org/10.1016/j.ygeno.2018.02.002
Hein, J., Schierup, M., & Wiuf, C. (2004). Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press, USA.
Ibeagha-Awemu, E. M., Peters, S. O., Akwanji, K. A., Imumorin, I. G., & Zhao, X. (2016). High density genome wide genotyping-by-sequencing and association identifies common and low frequency SNPs, and novel candidate genes influencing cow milk traits. Scientific reports, 6(1), 31109.
Jaglan, K., Ravikumar, D., Sukhija, N., George, L., Alex, R., Vohra, V., & Verma, A. (2023). Genomic clues of association between clinical mastitis and SNPs identified by ddRAD sequencing in Murrah buffaloes. Animal Biotechnology, 34(9), 4538-4546.
Kanaka, K. K., Sukhija, N., Goli, R. C., Singh, S., Ganguly, I., Dixit, S. P., ... & Malik, A. A. (2023). On the concepts and measures of diversity in the genomics era. Current Plant Biology, 33, 100278.
Lachance, J., & Tishkoff, S. A. (2013). SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. Bioessays, 35(9), 780-786.
Langmead, B., & Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nature methods, 9(4), 357-359.
Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27 (21):2987-2993. https://doi.org/10.1093/bioinformatics/btr509
Malik AA, Sharma R, Ahlawat S, Deb R, Negi MS, Tripathi SB (2018) Analysis of genetic relatedness among Indian cattle (Bos indicus) using genotyping-by-sequencing markers. Animal genetics 49 (3):242-245. https://doi.org/10.1111/age.12650
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., Hunter, D. J., ... & Visscher, P. M. (2009). Finding the missing heritability of complex diseases. Nature, 461(7265), 747–753.
Meena, D. C., Garai, S., Maiti, S., & Mandi, K. (2019). Pastoralists in Modern India: Current Status and Future Prospectus. In: Jayakumar R, Laddunuri M M, Kumar A, Duvvuru R R (eds) Research trends in multidisciplinary research, 1st edn. Akinik publications, New Delhi
Parameswari, S., Karthickeyan, S. M. K., Arunachalam, K., & Gopinathan, A. (2021). Phenotypic characterisation of Alambadi cattle-An unexplored indigenous population of south India. Indian J. Anim. Sci, 91(11), 925-930.
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLOS ONE 7 (5):e37135. https://doi.org/10.1371/journal.pone.0037135
Planning Commission of India (1989). Agro-climatic regional planning: An overview. Government of India, New Delhi
Rahman, J. U., Kumar, D., Singh, S. P., Shahi, B. N., Ghosh, A. K., Verma, M. K., ... & Sharma, R. K. (2023). Genome-wide identification and annotation of SNPs and their mapping in candidate genes related to milk production and fertility traits in Badri cattle. Tropical Animal Health and Production, 55(2), 117.
Raja, T. V., Alex, R., Singh, U., Kumar, S., Das, A. K., Sengar, G., & Singh, A. K. (2023). Genome wide mining of SNPs and INDELs through ddRAD sequencing in Sahiwal cattle. Animal Biotechnology, 1–15. https://doi.org/10.1080/10495398.2023.2200517
Said, S., Putra, W. P. B., Anwar, S., Agung, P. P., & Yuhani, H. (2017). Phenotypic, morphometric characterization and population structure of Pasundan cattle at West Java, Indonesia. Biodiversitas Journal of Biological Diversity, 18(4), Article 4. https://doi.org/10.13057/biodiv/d180443
Sambrook, J., & Russell, D. W. (2001). Molecular Cloning-Sambrook & Russel-Vol. 1, 2, 3. Cold Springs Harbor Lab Press: Long Island, New York.
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27 (6):863-864. https://doi.org/10.1093/bioinformatics/btr026
Selvan, A. S., Thangaraj, S., Dash, S., Karthikeyan, A., & Karthickeyan, S. (2022). Multivariate analysis of morphometric traits of Malaimadu cattle–Autochthonous draught cattle of south India. https://doi.org/10.21203/rs.3.rs-2191383/v1
Shabtay, A. (2015). Adaptive traits of indigenous cattle breeds: The Mediterranean Baladi as a case study. Meat science, 109, 27-39.
Sharma, R., Pandey, A. K., Singh, P. K., Maitra, A., Mukesh, M., Singh, S. R., & Singh, B. (2012). Characterization of Shahabadi cattle of Bihar—Phenotypic and molecular approaches. Indian Journal of Animal Sciences, 82(3), 318.
Sharma, R., Kishore, A., Mukesh, M., Ahlawat, S., Maitra, A., Pandey, A. K., & Tantia, M. S. (2015). Genetic diversity and relationship of Indian cattle inferred from microsatellite and mitochondrial DNA markers. BMC genetics, 16(1), 1-12.
Sukhija, N., Malik, A. A., Devadasan, J. M., Dash, A., Bidyalaxmi, K., Ravi Kumar, D., ... & Verma, A. (2023). Genome-wide selection signatures address trait specific candidate genes in cattle indigenous to arid regions of India. Animal Biotechnology, 1-15.
Sukhija, N., Goli, R. C., Sukhija, M., Rathi, P., Jaglan, K., Dash, A., ... & Kanaka, K. K. (2024a). Evolutionary stamps for adaptation traced in Cervus nippon genome using reduced representation sequencing. Conservation Genetics Resources, 16(1), 135-146.
Sukhija, N., Choudhary, A., Kanaka, K. K., Jaglan, K., Devadasan, M. J., Sivalingam, J., & Verma, A. (2024b). Genetic blueprinting of novel and performance traits-related SNPs in Indian Gir cattle using latest reference assembly: ddRAD analysis of Gir cattle for SNP discovery. Indian Journal of Dairy Science, 77(1).
Surati, U., Verma, A., & Niranjan, S. K. (2023). Genome-wide in silico analysis leads to identification of deleterious L290V mutation in RBBP5 gene in Bos indicus. Animal Biotechnology, 1–9. https://doi.org/10.1080/10495398.2023.2199502
Tantia, M. S., Vijh, R. K., Bhasin, V., Sikka, P., Vij, P. K., Kataria, R. S., Mishra, B. P., Yadav, S. P., Pandey, A. K., & Sethi, R. K. (2011). Whole-genome sequence assembly of the water buffalo (Bubalus bubalis). Indian J Anim Sci, 81(5), 38.
Tyagi, S. K., Mehrotra, A., Singh, A., Kumar, A., Dutt, T., Mishra, B. P., & Pandey, A. K. (2021). Comparative signatures of selection analyses identify loci under positive selection in the Murrah Buffalo of India. Frontiers in Genetics, 1915. https://doi.org/10.3389/fgene.2021.673697
Vineeth MR, Surya T, Sivalingam J, Kumar A, Niranjan SK, Dixit SP, Singh K, Tantia MS, Gupta ID (2020) Genome-wide discovery of SNPs in candidate genes related to production and fertility traits in Sahiwal cattle. Tropical Animal Health and Production 52 (4):1707-1715. https://doi.org/10.1007/s11250-019-02180-x
Vohra, V., Mishra, A., Niranjan, S., Chopra, A., Kumar, M., & Joshi, B. (2016). Phenotypic characterization, management and performance of Belahi cattle. The Indian Journal of Animal Sciences, 86, 355.
Wang W, Gan J, Fang D, Tang H, Wang H, Yi J, Fu M (2018) Genome-wide SNP discovery and evaluation of genetic diversity among six Chinese indigenous cattle breeds in Sichuan. PloS one 13 (8):e0201534-e0201534. https://doi.org/10.1371/journal.pone.0201534
Wara, A. B., Kumar, A., Singh, A., Arthikeyan, A. K., Dutt, T. R. I. V. E. N. I., & Mishra, B. P. (2019). Genome wide association study of test day’s and 305 days milk yield in crossbred cattle. Indian J. Anim. Sci, 89, 861-865.
Weldegerima, T. M. (2018). Characterization of productive and reproductive performances, morphometric and challenges and opportunities of indigenous cattle breeds of Ethiopia: A review. International Journal of Livestock Production, 9(3), 29–41. https://doi.org/10.5897/IJLP2017.0426
Acknowledgements
Authors would like to thank the Director of ICAR-National Bureau of Animal Genetics and Resources for granting funds and facilities to carry out research and analysis
Funding
This work was supported by ICAR-National Bureau of Animal Genetics and Resources (Grant number: 1.33).
Author information
Authors and Affiliations
Contributions
Jayakumar Sivalingam—Conceived the idea, carried out the work, data analysis and drating of the manuscript, S K Niranjan—Carried out the work, Dinesh Kumar Yadav—Helped in carrying out the analysis, S P Singh—Helped in the survey, Nidhi Sukhija—Drafting the manuscript, Kanaka K K—Drafting the manuscript, P K Singh—Conceived the idea and Ajit Pratap Singh—Helped in the survey and characterization of the animals.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
The blood samples of the animals were collected in accordance with the relevant guidelines and regulations as approved by IAEC (Institutional Animal Ethics Committee) of ICAR-National Bureau of Animal Genetics Resources, Haryana, India.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sivalingam, J., Niranjan, S.K., Yadav, D.K. et al. Phenotypic and genetic characterization of unexplored, potential cattle population of Madhya Pradesh. Trop Anim Health Prod 56, 102 (2024). https://doi.org/10.1007/s11250-024-03946-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-024-03946-8