Abstract
Conservation of locally adapted indigenous livestock breeds has become an important objective in sustainable animal breeding, as these breeds represent a unique genetic resource. Therefore, the Agricultural Research Council of South Africa initiated a conservation programme for four South African indigenous chicken breeds. The evaluation and monitoring of the genetic constitution of these conservation flocks is important for proper management of the conservation programme. Using molecular genetic analyses, the effective population sizes and relatedness of these conservation flocks were compared to village (field) chicken populations from which they were derived. Genetic diversity within and between these populations are further discussed within the context of population size. The conservation flocks for the respective breeds had relatively small effective population sizes (point estimate range 38.6–78.6) in comparison to the field populations (point estimate range 118.9–580.0). Furthermore, evidence supports a transient heterozygous excess, generally associated with the occurrence of a recent population bottleneck. Genetic diversity, as measured by the number of alleles, heterozygosity and information index, was also significantly reduced in the conservation flocks. The average relatedness amongst the conservation flocks was high, whilst it remained low for the field populations. There was also significant evidence for population differentiation between field and conservation populations. F st estimates for conservation flocks were moderate to high with a maximum reached between VD_C and VD_F (0.285). However, F st estimates for field population were excessively low between the NN_C and EC_F (0.007) and between EC_F and OV_F (0.009). The significant population differentiation of the conservation flocks from their geographically correlated field populations of origin is further supported by the analysis of molecular variance (AMOVA), with 10.51 % of genetic diversity ascribed to population differences within groups (F SC = 0.106). The results suggest that significant genetic erosion has occurred within the conservation flocks due to inbreeding, pronounced effects of random drift and selection. It might be necessary to introduce new breeding individuals from the respective field populations in order to increase the effective population sizes of the conservation flocks and counter the effects of genetic erosion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, there have been particular concerns raised regarding the adaptability and evolutionary potential of highly industrialised livestock breeds, considering global climate change and food security (Ajmone-Marsan and The GLOBALDIV Consortium 2010; Groeneveld et al. 2010). Indigenous livestock breeds generally possess adaptive characteristics that make them better suited to local environmental (often harsh) conditions. These breeds represent a unique genetic resource for long-term and sustainable animal genetic improvement (Taberlet et al. 2008; Medugorac et al. 2009). However, many indigenous breeds remain poorly characterised and are currently threatened by extinction due to changing production systems, preferring exotic commercial breeds and indiscriminate crossbreeding (Besbes 2009). To prevent the irreversible erosion of animal genetic resources that might compromise future breeding programmes, the FAO initiated the “Global Plan of Action for Animal Genetic Resources” to facilitate the characterisation and conservation of indigenous livestock breeds (FAO 2007a).
Chicken genetic resources are probably the most endangered and under-conserved of all livestock species, with approximately 33 % of the world’s chicken breeds considered endangered (FAO 2007b; Hoffmann 2009). In South Africa and most African countries, indigenous chickens are raised by smallholder farmers with little resources and are considered important genetic resources that should be conserved against production threats and replacement with commercial hybrids (Muchadeyi et al. 2007). Characterisation of these genetic resources will serve as an essential prerequisite for the identification and effective management and utilisation of South African indigenous chickens, which will facilitate their conservation. Ruane (1999) further confirmed that adaptive features, traits of scientific and economic interest, cultural-historical values, strong links to regional traditions and ability to generate income associated with most of these village chicken populations further justify conservation efforts. For this reason, phenotypic observations or monitoring of productive traits combined with molecular analysis can be useful information for conservation decisions.
The importance of conservation of chicken genetic resources has long been recognised in South Africa (ARC 2006). The Animal Production Institute of the Agricultural Research Council initiated a breeding programme in 1994 for four indigenous chicken breeds as a base for conservation and genetic improvement. The four breeds that form part of the conservation flocks include the Venda, Naked Neck, Ovambo and Potchefstroom Koekoek. There are no particular breed standards, and the breeds are generally classified based on geographic region of origin and broad morphological characteristics. Little is known about the origin and history of these breeds with the exception of the Potchefstroom Koekoek (for which breed standards have been established) that was developed as a dual purpose breed during the 1940s/1950s through crosses between the White Leghorn, Black Austrolorp and Plymouth Rock breeds (Van Marle-Köster and Nel 2000).
Conservation decisions of South African indigenous chickens were mainly based on general population trends in reproduction and production parameters with little considerations for genetic factors affecting extinction probability. This oversight might jeopardise the preservation of these chicken genetic resources in the long term. Consequently, initial studies were conducted to evaluate the extent of intra- and interbreed genetic diversity (as measured by microsatellite loci) using the resource flocks held for conservation. Van Marle-Köster et al. (2008) reported moderate genetic diversity within the respective breeds (He = 0.50–0.65), as well as moderately high F is estimates (between 0.21 and 0.35), suggesting possible inbreeding. On the contrary, Mtileni et al. (2012) reported considerably low F is estimates, ranging between −0.048 and 0.041, with heterozygosity estimates being comparable (H e = 0.51–0.62) between the two studies. Both studies also supported significant population substructure between the various conservation flocks, but this was more evident in the study by Mtileni et al. (2012).
To formulate appropriate managerial strategies for these indigenous chickens and to prevent genetic erosion, it is necessary to assess the causality of such discrepancies. Effective population size has been used as a criterion for determining the extinction risk and for setting conservation limits (CLs) of single populations (and/or species), e.g. in international guidelines for categorising threatened species (Mace and Lande 1991). An analysis of the effective population size of the conserved and the village chicken population will have a major impact on the dynamics of these two population categories and will also help in quantifying whether these two particular populations are affected by drift or inbreeding. The effective population size (Ne) determines the degree to which gene frequencies are faithfully transmitted across generations (Wright 1931), and it is a key factor in the nearly neutral theory of molecular evolution, because the fate of a mutation is determined by the product N es. Furthermore, this effective population size analysis will also take into account not only the current census size of a population but also the history of the population. This study, therefore, aimed to evaluate the intra- and interpopulation genetic diversity of these flocks within the context of demographic dynamics [effective population size (N e), population contractions or expansions] and inbreeding, in particular, the possibility of a recent population bottleneck, creating a transient heterozygous excess, deflating the F is estimates and consequently leading to under-estimation of true levels of current inbreeding.
Materials and methods
Population representative samples were collected from four indigenous chicken breeds, kept for conservation purposes at the Poultry Breeding Resource Unit of the Agricultural Research Council: Venda (VD_C, n = 30), Ovambo (OV_C, n = 26), Naked Neck (NN_C, n = 29) and Potchefstroom Koekoek (PK_C, n = 29). For comparison, a random sample of village chickens from three South African provincial regions was also taken: Venda (Limpopo Province, VD_F, n = 30), Ovambo (Northern Cape Province, OV_F, n = 42) and the Eastern Cape Province (EC_F, n = 26). Village chicken populations were sampled from farming areas from which the current conservation flocks originated, which are Limpopo Province (VD_F chickens) and Northern Cape Province along the border with Namibia (OV_F chickens) as well as the Eastern Cape Province (EC_F chickens). Ninety-eight households were randomly selected from 23 villages of Vhembe and Mopani Districts in the Limpopo Province, Kgalagadi and Namaqua Districts of the Northern Cape Province and Alfred Nzo and OR Tambo Districts of the Eastern Cape Province. For each district, 2–5 villages were selected. The distance between villages within district ranged from 20 to 40 km, 100 to 500 km between districts within a province and over 1000 km between provinces. One chicken was sampled per household. Blood was collected from the wing vein onto FTA micro-cards (Whatman Bio Science, Kent, UK) for each individual. A standard phenol/chloroform extraction protocol was followed for DNA isolation (Sambrook and Russell 2001). Each individual animal was genotyped for a set of 29 autosomal microsatellite markers as previously described in Mtileni et al. (2012).
Micro-checker v.2.2.3 (Van Oosterhout et al. 2004) was used to detect genotyping errors due to allelic dropout, stuttering and null alleles (null allele estimates as per the method of Brookfield 1996). Departures from Hardy-Weinberg equilibrium were evaluated by means of the exact probability test (500 batches, 10,000 iterations) in Genepop v.4.0 (Rousset 2008). Markers were also tested for neutrality using an F st outlier test as implemented in Lositan v.1.44 (10,000 permutations assuming the infinite alleles model, with a correction for false discovery rate at 0.01 and statistical significance at the 5 % nominal level) (Antao et al. 2008). The following genetic diversity estimates were calculated in Genalex v.6.4 (Peakall and Smouse 2006): Observed (Ho) and expected heterozygosity (He), number of alleles (An), effective number of alleles (A e) and the information (Shannon-Weaver) index (I). To test whether there were significant differences in diversity between the village (field) chickens and the conservation flocks a Kruskal-Wallis test was performed (nominal level of 5 % for statistical significance) in XLStatistics v.10.05.03 (Carr 2010). Pairwise Fst estimates (significance testing: 10,000 permutations, at a nominal level of 5 %) and a locus-by-locus hierarchical analysis of molecular variance (AMOVA) (significance testing: 10,000 permutations, at a nominal level of 5 %) was calculated in Arlequin v.3.5.1.2 (Excoffier and Lischer 2010). Where appropriate, the conservation flocks were grouped with the relevant geographically correlated field populations for the hierarchical AMOVA. To further elucidate the relationship between the various populations a dendrogram was constructed using Nei’s genetic distance, Da (Nei 1978) and the neighbour joining clustering method (significance testing: 1000 bootstrap replicates) in Treefit v.1.0 (Kalinowski 2009). A principle coordinate analysis (PCoA) was also conducted per population and sample in Genalex. Effective population sizes were calculated using the heterozygous excess test in NeEstimator v.1.3 (Peel et al. 2004) as well as the linkage disequilibrium (LD) test (minimum allele frequency, 0.02) in LDNe v.1.0 (Waples 2006). The occurrence of recent bottlenecks was evaluated by means of the Wilcoxon signed rank test [assuming the infinite alleles (IAM) and the two-phase mutation models (TPM), 10,000 replicates at 5 % nominal level] and the mode-shift test, in Bottleneck v.1.2.02 (Piry et al. 1999). Furthermore, to evaluate the extent of inbreeding, mean relatedness was calculated for each population using the method of Queller and Goodnight (1989) (significance testing by 1000 bootstrap replicates) as well as mean F is estimated in Genalex.
Results and discussion
The results are discussed in light of conservation genetic theory and empirical results on the fitness consequences of loss of genetic variation in conservation flock and field population within the context of demographic dynamics [effective population size (N e), population contractions or expansions] and inbreeding, in particular, the possibility of a recent population bottleneck, creating a transient heterozygous excess, deflating the F is estimates and consequently leading to under-estimation of true levels of current inbreeding. In general, tables and figures summarise the analysed data. There was no evidence for large allele dropout or excessive stuttering that could have influenced allele scoring. However, eight loci demonstrated genotypic patterns consistent with the presence of null alleles (null allele frequencies ranged between 0.07 and 0.13; P < 0.05) in one or more populations, and these loci were removed from further analyses. The remaining 21 loci are all in Hardy-Weinberg expectations (P > 0.01), and none were identified as outliers in the F st outlier test and were thus presumed selectively neutral. Estimates of genetic diversity were low to moderate across the populations. The VD_C population consistently demonstrated the lowest genetic diversity across all measures (H o, H e, A n, A e, I), whilst the OV_F and EC_F populations showed the highest levels of genetic diversity (Fig. 1). In general, the conservation flocks displayed significantly lower genetic diversity across all measures when compared to the field populations (Kruskal-Wallis test, P > 0.01). The low genetic diversity in these conservation flocks was previously confirmed (Van Marle-Köster et al. 2008; Mtileni et al. 2012) and has also been reported for other chicken breeds under conservation (Dávila et al. 2009).
Despite the loss in genetic diversity, the conservation flocks demonstrated lower F is estimates than the field populations, even demonstrating significant heterozygous excess for the NN_C population (Fig. 2a). On the contrary, estimates for average relatedness (r) amongst the conservation flocks were significantly higher than for the field populations, suggesting considerable inbreeding (Fig. 2b). The heterozygosity excess estimate for N e suggests fairly large N e for all populations with the exception of the NN_C and VD_C populations. The LD estimate for N e seems to demonstrate more realistic estimates, comparable to what would be expected: generally larger N e for the field populations (point estimate range 118.9–580.0), with smaller estimates for the conservation flocks (point estimate range 38.6–78.6). Both methods of estimation showed that the NN_C and VD_C populations had the smallest N e (Table 1). The LD estimate for N e is probably a more reliable estimate given the limited sample size and heterozygosity excess due to a possible bottleneck event (Waples and Do 2010). Under a strict infinite allele model, the Wilcoxon signed rank test suggested that recent population bottlenecks have occurred for all populations; however, under the two-phase model only, one of the field populations (EC_F) and two of the conservation flock populations (OV_C and NN_C) demonstrated significant evidence. This evidence for recent bottlenecks was not mirrored by the mode-shift test (Table 1). When population size is small, genetic drift may outweigh the force of selection, leading to the loss of adaptive genetic variation and the fixation of deleterious alleles (Kimura and Crow 1963). Evidence showing that selection efficiency and effective population size are positively correlated is increasing in the last years. Lynch and Conery (2003) have proposed that the changes in genome complexity from prokaryotes to multicellular eukaryotes, including gene number, intron abundance and mobile genetic elements, emerged passively in response to long-term population-size reductions.
The fairly low estimates for F is for the conservation flocks contradict the findings of Van Marle-Köster et al. (2008), where estimates larger by a factor of ten were reported, although the general diversity statistics were comparable. F is is a function of heterozygosity (indirectly homozygosity), which in turn is often assumed to be a function of inbreeding (Szulkin et al. 2010). Although this is generally the case for small isolated populations (e.g. Ruiz-López et al. 2012), if a population has undergone a recent bottleneck, the loss in allelic diversity is not accompanied by an immediate decrease in heterozygosity. This creates a transient heterozygosity excess (Cornuet and Luikart 1997; Luikart and Cornuet 1998) and, therefore, the direct correlation between heterozygosity and inbreeding is lost. The relatively high estimates for relatedness amongst the conservation indicate considerable inbreeding in comparison to estimates for the field populations (Table 1). Thus, the low F is estimates are most likely a consequence of recent bottleneck in the conservation flocks as supported by the Wilcoxon signed rank test. There is further evidence that the effective population sizes of the respective conservation flocks are small and have been reduced: the LD estimate for N e clearly shows a trend of smaller N e for the conservation flocks in comparison to the field populations. Current analysis of N e revealed that at least three of the conservation flocks may be under threat as evident by their N e less than the critical minimum value (N e ≈ 50), which is generally required for maintaining a viable population (Taberlet et al. 2008). There is some indication that a recent bottleneck also occurred in the field populations; this may reflect a decline in the maintenance of livestock due to urbanisation but needs further investigation. Thus, our results support the hypothesis that species with lower polymorphism have larger content of repetitive sequences in their genomes, suggesting a diminished efficiency of selection in species with smaller effective population size, whilst species with larger effective population size undergo higher levels of adaptive selection (Petit and Barbadilla 2008).
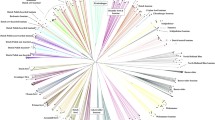
The genetic differentiation within the field populations was low to moderate, with the highest pairwise F st = 0.101 estimate observed between VD_F and EC_F (Table 2). This low to moderate differentiation was further supported by the PCoA, showing a close clustering of the field populations (Fig. 3) with considerable overlap in the per sample analysis (Figure S1) and comparatively low bootstrap values at branching points in the dendrogram (Fig. 4). Previously, using a Bayesian clustering approach, it was concluded that there was little evidence for population structure between the village chickens in South Africa (Mtileni et al. 2012). However, the current analyses suggest that there might be slight differentiation between populations from various geographic regions. In pairwise comparisons including conservation flocks, F st estimates were moderate to high with a maximum reached between VD_C and VD_F (0.285). However, pairwise F st estimates between the NN_C and EC_F and between EC_F and OV_F were excessively low (0.007 and 0.009, respectively) (Table 2). Nonetheless, cluster analysis supported the genetic distinctness of the NN_C population (Figs. 3 and 4). The significant population differentiation of the conservation flocks from their geographically correlated field populations of origin is further supported by the AMOVA, with 10.51 % of genetic diversity ascribed to population differences within groups (F SC = 0.106) (Table 3) and the divergent population clusters, for conservation and field populations, in the dendrogram. The genetic distinctness of the conservation flocks has previously been confirmed (Van Marle-Köster et al. 2008; Mtileni et al. 2012). The variation amongst groups (F CT = 0.006) failed to reach statistical significance probably due to the low degree of population differentiation of the field populations (Table 3). The long branch lengths as observed for the conservation flocks on the dendrogram are also often indicative of small and isolated populations (Medugorac et al. 2009).
Dendrogram based on D a and constructed using the neighbour joining clustering method. Nodal values—bootstrap percentage based on 1000 replicates; R 2 value—a measure of how well the current tree confirmation represents the observed genetic variation (Kalinowski 2009)
In conclusion, there is sufficient evidence to suggest that the current conservation flocks kept at the Poultry Breeding Resource Unit of the Agricultural Research Council (South Africa) are genetically distinct from the village chickens from where they were sourced originally. These conservation flocks represent small and isolated populations with relatively high levels of inbreeding and genetic erosion. Results from the current study raise some concerns with regard to the long-term sustainability of the conservation programme. Furthermore, there is evidence suggesting that the conservation flocks have undergone a recent bottleneck that is not associated with the original founder event in 1994. This bottleneck could be due to selective breeding to enhance the productivity of these indigenous breeds. It may, therefore, be advisable to maintain separate, but connected programmes for conservation and genetic improvement. As such, the conservation programme may feed into the genetic improvement programme, but preserving genetic diversity within the conservation nucleus populations must be prioritised in order to ensure population adaptability and resilience in the aim of future breeding objectives. Therefore, it may be advantageous to augment the current conservation flocks with individual animals from geographically correlated populations in an effort to restore lost genetic diversity and negate the adverse effects of small effective population sizes and inbreeding.
References
Ajmone-Marsan, P. and The GLOBALDIV Consortium, 2010. A global view of livestock biodiversity and conservation—GLOBALDIV. Animal Genetics, 41 (Suppl. 1), 1–5.
Antao, T., Lopes, A., Lopes, R..L., Beja-Pereira, A. and Luikart, G., 2008. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outliner method. BMC Bioinformatics, 9, 323.
ARC (2006) Genetic characterization and distances of indigenous livestock breeds. Accessed February 11, 2010, from: http://www.arc.agric.za/home.asp?pid=567.
Besbes, B., 2009. Genotype evaluation and breeding of poultry for performance under sub-optimal village conditions. World’s Poultry Science Journal, 65, 260–271.
Brookfield, J.F.Y., 1996. A simple new method for estimating null allele frequency from heterozygote deficiency. Molecular Ecology, 5, 453–455.
Carr, R., 2010. XLStatistics 10.05.30, XLent Works, Australia.
Cornuet, J.M. and Luikart, G., (1997) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144, 2001–2014.
Dávila, S.G., Gil, M.G., Resino-Talaván, P. and Campo J.L., 2009 Evaluation of diversity between different Spanish chicken breeds, a tester line, and a White Leghorn population based on microsatellite markers. Poultry Science, 88, 2518–2525.
Excoffier L. and Lischer H.E.L., 2010. Arlequin suite version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecological Resources, 10, 564–567.
FAO., 2007a. The Global Plan of Action for Animal Genetic Resources and the Interlaken Declaration on Animal Genetic Resources. FAO, Rome, Italy.
FAO., 2007b. The State of the World’s Animal Genetic Resources for Food and Agriculture. In: Rischkowsky B., Pilling D. (Eds.). FAO, Rome.
Groeneveld, L.F., Lenstra, J.A., Eding, H., Toro, M.A., Scherf, B., Pilling, D., Negrini, R., Finlay, E.K., Jianlin, H., Groeneveld, E., Weigend, S. and The GLOBALDIV Consortium., 2010. Genetic diversity in farm animals - a review. Animal Genetics, 41 (Suppl. 1), 6–31.
Hoffmann, I., 2009. The global plan of action for animal genetic resources and the conservation of poultry genetic resources. World’s Poultry Science Journal, 65, 286–297.
Kalinowski, S.T., 2009. How well do evolutionary trees describe genetic relationships among populations? Heredity, 102, 506–513.
Kimura, M. & Crow, J. F. 1963. The measurement of effective population number. Evolution 17, 279–288.
Lynch, M. and Conery, J. S. 2003 The origins of genome complexity. Science 302, 1401–1404. (doi:10.1126/science.1089370)
Luikart, G. and Cornuet, J.M., 1998. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology, 12, 228–237.
Mace, G. M., and Lande, R. 1991. Assessing extinction threats: towards a reassessment of IUCN endangered species categories. Conservation Biology, 5: 148e157.
Medugorac, I., Medugorac, A., Russ, I., Veit-Kensc, C.E., Taberlet, H.P., Luntz, B., Mix, H.M. and Forster, M., 2009. Genetic diversity of European cattle breeds highlights the conservation value of traditional unselected breeds with high effective population size. Molecular Ecology, 18, 3394–3410.
Mtileni, B.J., Muchadeyi, F.C., Maiwashe, A., Groeneveld, E., Groeneveld, L.F., Dzama, K. and Weigend, S., 2012. Genetic diversity and conservation of South African indigenous chicken populations. Journal of Animal Breeding and Genetics, 128, 209–218.
Muchadeyi, F.C., Wollny, C.B.A., Eding, H., Weigend S., Makuza, S.M., Simianer, H., (2007). Variation in village chicken production systems among agro-ecological zones of Zimbabwe. Trop. Anim. Health. Prod., 39:453–461.
Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583–590.
Peakall, R. and Smouse, P.E., 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecological Notes, 6, 288–295.
Peel, D., Ovenden, J.R. and Peel, S.L., 2004. NeEstimator: software for estimating effective population size, Version 1.3. Queensland Government, Department of Primary Industries and Fisheries.
Petit, N. & Barbadilla, A. 2008. Selection efficiency and effective population size in Drosophila species. J. Evol. Biol. 22, 515–526. (doi:10.1111/j.1420-9101.2008.01672.x).
Piry, S., Luikart, G. and Cornuet, J.M., 1999. BOTTLENECK: a computer program for detecting recent reductions in effective population size using allele frequency data. Journal of Heredity, 90, 502–503.
Queller, D.C. and Goodnight, K.F., 1989. Estimating relatedness using genetic markers. Evolution, 43, 258–275.
Rousset, F., 2008. GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecological Resources, 8, 103–106.
Ruane, J. 1999. Genebanks and the Conservation of Farm Animal Genetic Resources. In: Oldenbroek, J.K. (ed.), DLO Institute for Animal Science and Health, Lelystad, the Netherlands, pp. 59–73.
Ruiz-lópez, M., Gãnan, N., Godoy, J.A., Del Olmo, A., Garde, J., Espeso, G., Vargas, A., Martinez, F., Roldán, E.R.S. and Gomendio, M., 2012. Heterozygosity-fitness correlations and inbreeding depression in two critically endangered mammals. Conservation Biology, 26, 1121–1129.
Sambrook, J. and Russell, D.W., 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, New York, USA.
Szulkin, M., Bierne, N. and David P., 2010. Heterozygosity-fitness correlations: a time for reappraisal. Evolution, 64, 1202–1217.
Taberlet, P., Valentini, A., Rezaei, H.R., Naderi, S., Pompanon, F., Negrini, R. and Ajmone-Marsan, P., 2008. Are cattle, sheep, and goats endangered species? Molecular Ecology, 17, 275–284.
Van Marle-Köster, E., Hefer, C.A., Nel, L.H. and Groenen, M.A.M., 2008. Genetic diversity and population structure of locally adapted South African chicken lines: Implications for conservation. South African Journal of Animal Science, 38, 271–281.
Van Marle-Köster, E. and Nel, L.H., 2000. Genetic characterization of native southern African chicken populations: evaluation and selection of polymorphic microsatellite markers. South African Journal of Animal Science, 30, 1–6.
Van Oosterhout C., Hutchinson W.F., Wills D.P.M. and Shipley P., 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 545–538.
Waples, R.S. and Do, C., 2010. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evolutionary Applications, 3, 244–262.
Waples, R.S. 2006. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conservation Genetics, 7, 167–184.
Wright, S. 1931. Evolution in Mendelian populations. Genetics 16, 97–159.
Acknowledgments
The Department of Science and Technology in South Africa is acknowledged for the financial support of this study. We also thank the farmers in the three provinces of South Africa for their cooperation during blood collection.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 103 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mtileni, B., Dzama, K., Nephawe, K. et al. Estimates of effective population size and inbreeding in South African indigenous chicken populations: implications for the conservation of unique genetic resources. Trop Anim Health Prod 48, 943–950 (2016). https://doi.org/10.1007/s11250-016-1030-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-016-1030-9