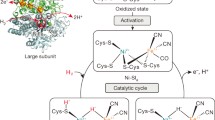
Abstract
X-ray crystallography is the most powerful tool for obtaining structural information about protein molecules, affording accurate and precise positions for all of the atoms in the protein except for hydrogen. However, hydrogen species play crucial roles in the physiological functions of enzymes, including molecular recognition through hydrogen bonding and catalytic reactions involving proton transfer. Neutron crystallography enables direct identification of the positions of hydrogen species. [NiFe]-hydrogenase from Desulfovibrio vulgaris Miyazaki F is an enzyme that catalyzes the reversible oxidation of molecular hydrogen. It contains a bimetallic Ni–Fe active site for the catalytic reaction and three Fe–S clusters for electron transfer. Previous X-ray structure analyses of the enzyme under various oxidation conditions have revealed that the active site changes its coordination structure depending on the redox state. In the inactive air-oxidized form, an oxygen species was identified between the Ni and Fe atoms, whereas in the active H2-reduced form, subatomic-resolution X-ray structure analysis and single-crystal EPR analyses indicated a hydride ligand between the two metal atoms. However, the assignment of the hydride moiety by X-ray crystallography remains controversial, and the proton transfer pathways in the molecule are still ambiguous. To allow neutron diffraction experiments, large crystals of [NiFe]-hydrogenase were prepared by the vapor diffusion method with the macroseeding technique according to the two-dimensional phase diagram (protein concentration vs. precipitant concentration). Neutron diffraction data were collected at approximately 2.0 Å resolution at cryogenic temperature using a gas-stream cooling system to trap short-lived intermediates in the catalytic reaction.






Similar content being viewed by others
References
Hazemann I, Dauvergne MT, Blakeley MP, Meilleur F, Haertlein M, Van Dorsselaer A, Mitschler A, Myles DAA, Podjarny A (2005) Acta Cryst D61:1413–1417
Coates L, Cao HB, Chakoumakos BC, Frontzek MD, Hoffmann C, Kovalevsky AY, Liu Y, Meilleur F, dos Santos AM, Myles DAA, Wang XP, Ye F (2018) Rev Sci Instrum 89:092802
Kwon H, Langan PS, Coates L, Raven EL, Moody PCE (2018) Acta Cryst D74:792–799
Blakeley MP, Cianci M, Helliwell JR, Rizkallah PJ (2004) Chem Soc Rev 33:548–557
Vignais PM, Billoud B, Meyer J (2001) FEMS Microbiol Rev 25:455–501
Ogata H, Nishikawa K, Lubitz W (2015) Nature 520:571–574
Vignais PM, Billoud B (2007) Chem Rev 107:4206–4272
Stiebritz MT, Reiher M (2012) Chem Sci 3:1739–1751
Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE (2016) ISME J 10:761–777
Haumann M, Porthun A, Buhrke T, Liebisch P, Meyer-Klaucke W, Friedrich B, Dau H (2003) Biochemistry 42:11004–11015
Lubitz W, Ogata H, Rüdiger O, Reijerse E (2014) Chem Rev 114:4081–4148
Higuchi Y, Yagi T, Yasuoka N (1997) Structure 5:1671–1680
Higuchi Y, Ogata H, Miki K, Yasuoka N, Yagi T (1999) Structure 7:549–556
Ogata H, Mizoguchi Y, Mizuno N, Miki K, Adachi S, Yasuoka N, Yagi T, Yamauchi O, Hirota S, Higuchi Y (2002) J Am Chem Soc 124:11628–11635
Fernandez VM, Hatchikian EC, Cammack R (1985) BBA-Prot Struct Mol Enzymol 832:69–79
Volbeda A, Martin L, Cavazza C, Matho M, Faber BW, Roseboom W, Albracht SPJ, Garcin E, Rousset M, Fontecilla-Camps JC (2005) J Biol Inorg Chem 10:239–249
Ogata H, Hirota S, Nakahara A, Komori H, Shibata N, Kato T, Kano K, Higuchi Y (2005) Structure 13:1635–1642
Barondeau DP, Roberts LM, Lindahl PA (1994) J Am Chem Soc 116:3442–3448
de Lacey AL, Hatchikian EC, Volbeda A, Frey M, Fontecilla-Camps JC, Fernandez VM (1997) J Am Chem Soc 119:7181–7189
Lamle SE, Albracht SP, Armstrong FA (2004) J Am Chem Soc 126:14899–14909
Ogata H, Kellers P, Lubitz W (2010) J Mol Biol 402:428–444
Barilone JL, Ogata H, Lubitz W, van Gastel M (2015) Phys Chem Chem Phys 17:16204–16212
Tai H, Nishikawa K, Higuchi Y, Mao ZW, Hirota S (2019) Angew Chem 131:13419–13424
Evans RM, Brooke EJ, Wehlin SA, Nomerotskaia E, Sargent F, Carr SB, Phillips SEV, Armstrong (2016) FA Nat Chem Biol 12:46–50
Brecht M, van Gastel M, Buhrke T, Friedrich B, Lubitz W (2003) J Am Chem Soc 125:13075–13083
Foerster S, Van Gastel M, Brecht M, Lubitz W (2005) J Biol Inorg Chem 10:51–62
Haas C, Drenth J (1999) J Cryst Growth 196:388–394
Saridakis E, Chayen NE (2000) Protein Sci 9:755–757
George A, Wilson WW (1994) Acta Cryst D50:361 – 365
Hussein R, Ibrahim M, Chatterjee R, Coates L, Müh F, Yachandra VK, Yano J, Kern J, Dobbek H, Zouni A (2018) Cryst Growth Des 18:85–94
Budayova-Spano M, Koruza K, Fisher Z (2020) Methods Enzymol 634:21–46
Nishikawa K, Higuchi Y (2017) Int J Microgravity Sci Appl 34:340100
Yagi T, Kimura K, Daidoji H, Sakai F, Tamura S, Inouchi H (1976) J Biochem 79:661–671
Nakamura T, Kawasaki T, Hosoya T, Toh K, Ebine M, Birumachi A, Sakasai K, Soyama K, Katagiri M (2012) J Instrum 7:C02003
Tanaka I, Kusaka K, Hosoya T, Niimura N, Ohhara T, Kurihara K, Yamada T, Ohnishi Y, Tomoyori K, Yokoyama T (2010) Acta Cryst D66:1194–1197
Kusaka K, Hosoya T, Yamada T, Tomoyori K, Ohhara T, Katagiri M, Kurihara K, Tanaka I, Niimura N (2013) J Synchrotron Rad 20:994–998
Ohhara T, Kusaka K, Hosoya T, Kurihara K, Tomoyori K, Niimura N, Tanaka I, Suzuki J, Nakatani T, Otomo T, Matsuoka S, Tomita K, Nishimaki Y, Ajima T, Ryufuku S (2009) Nucl Instrum Methods Phys Res A 600:195–197
Yano N, Yamada T, Hosoya T, Ohhara T, Tanaka I, Niimura N, Kusaka K (2018) Acta Cryst D74:1041–1052
Afonine PV, Mustyakimov M, Grosse-Kunstleve RW, Moriarty NW, Langan P, Adams PD (2010) Acta Cryst D66:1153–1163
Kabsch W (2010) Acta Cryst D66:125–132
Hiromoto T, Nishikawa K, Inoue S, Matsuura H, Hirano Y, Kurihara K, Kusaka K, Cuneo M, Coates L, Tamada T, Higuchi Y (2020) Acta Cryst D76:946–953
Harrison K, Wu Z, Juers DH (2019) J Appl Cryst 52:1222–1232
Myles DAA, Dauvergne F, Blakeley MP, Meilleur F (2012) J Appl Cryst 45:686–692
Coates L, Tomanicek S, Schrader TE, Weiss KL, Ng JD, Jüttner P, Ostermann A (2014) J Appl Cryst 47:1431–1434
Coates L, Cuneo MJ, Frost MJ, He J, Weiss KL, Tomanicek SJ, McFeeters H, Vandavasi VG, Langan P, Iverson EB (2015) J Appl Cryst 48:1302–1306
Teixeira VH, Soares CM, Baptista AM (2008) Proteins 70:1010–1022
Ash PA, Hidalgo R, Vincent KA (2017) ACS Catal 7:2471–2485
Nishikawa K, Ogata H, Higuchi Y (2020) Chem Lett 49:164–173
Tai H, Higuchi Y, Hirota S (2018) Dalton Trans 47(13):4408–4423
Ilina Y, Lorent C, Katz S, Jeoung JH, Shima S, Horch M, Zebger I, Dobbek H (2019) Angew Chem Int Ed 58(51):18710–18714
Tai H, Nishikawa K, Inoue S, Higuchi Y, Hirota S (2015) J Phys Chem B 119:13668–13674
Acknowledgements
This study was partly supported by MEXT KAKENHI Grants-in-Aid for Scientific Research on Innovative Areas (Hydrogenomics) 18H05516 (to Y.H.) and for Scientific Research (A) 19H00984 (to Y.H.), (B) 19H03173 (to T.T.,) and (C) 16K07283 (to T.T.); the Hyogo Science and Technology Association (to T.H.); and a JST CREST grant JPMJCR12M4 (to Y.H.). Neutron diffraction experiments using iBIX of J-PARC were performed under user programs (proposal nos. 2014B0312, 2015A0159, 2016A0100, 2017A0036, 2017B0003, and 2018A0042) and the project for the Ibaraki prefectural local government beamline (proposal nos. 2019PX2003 and 2019PX2012). The research at ORNL’s SNS (IPTS nos. 19172.1 and 20933.1) was supported by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy. We thank Drs. K. Kusaka, L. Coates, M. Cuneo, Y. Hirano, K. Kurihara, and H. Matsuura for neutron diffraction experiments, and S. Inoue, K. Hataguchi, K. Matsumoto, Y. Ikeda, Y. Yamada, and J. Hiroki for technical assistance in this study.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hiromoto, T., Nishikawa, K., Tamada, T. et al. The Challenge of Visualizing the Bridging Hydride at the Active Site and Proton Network of [NiFe]-Hydrogenase by Neutron Crystallography. Top Catal 64, 622–630 (2021). https://doi.org/10.1007/s11244-021-01417-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11244-021-01417-0