Abstract
This paper focuses on the analysis of spatially correlated functional data. We propose a parametric model for spatial correlation and the between-curve correlation is modeled by correlating functional principal component scores of the functional data. Additionally, in the sparse observation framework, we propose a novel approach of spatial principal analysis by conditional expectation to explicitly estimate spatial correlations and reconstruct individual curves. Assuming spatial stationarity, empirical spatial correlations are calculated as the ratio of eigenvalues of the smoothed covariance surface Cov\((X_i(s),X_i(t))\) and cross-covariance surface Cov\((X_i(s), X_j(t))\) at locations indexed by i and j. Then a anisotropy Matérn spatial correlation model is fitted to empirical correlations. Finally, principal component scores are estimated to reconstruct the sparsely observed curves. This framework can naturally accommodate arbitrary covariance structures, but there is an enormous reduction in computation if one can assume the separability of temporal and spatial components. We demonstrate the consistency of our estimates and propose hypothesis tests to examine the separability as well as the isotropy effect of spatial correlation. Using simulation studies, we show that these methods have some clear advantages over existing methods of curve reconstruction and estimation of model parameters.
Similar content being viewed by others
1 Introduction
Functional data analysis (FDA) focuses on data that are infinite dimensional, such as curves, shapes, and images. Generically, functional data are measured over a continuum across multiple subjects. In practice, many data such as growth curves of different people, gene expression profiles, vegetation index across multiple locations, and vertical profiles of atmospheric radiation recorded at different times could naturally be modeled by FDA framework.
Functional data are usually modeled as noise-corrupted observations from a collection of trajectories that are assumed to be realizations of a smooth random function of time X(t), with unknown mean shape \(\mu (t)\) and covariance function Cov\((X(s),X(t)) = G(s,t)\). Functional principal components (fPCs) which are the eigenfunctions of the kernel G(s, t) provide a comprehensive basis for representing the data and hence are very useful in problems related to model building and prediction of functional data.
Let \({\phi _k(t),\ k = 1,2,\ldots ,K}\) and \({\lambda _k,\ k = 1,2,\ldots ,K}\) be the first K eigenfunctions and eigenvalues of G(s, t). Then
where \(\xi _{ik}\) are fPC scores which have mean zero and variance \(\lambda _k\). According to this model, all curves share the same mode of variations, \(\phi _k(t)\), around the common mean process \(\mu (t)\).
A majority of work in FDA assumes that the realizations of the underlying smooth random function are independent. There exists an extensive literature on functional principal components analysis (fPCA) for this case. For data observed at irregular grids, Yao et al. (2003) and Yao and Lee (2006) used local linear smoother to estimate the covariance kernel and integration method to compute fPC scores. However, the integration approximates poorly with sparse data. James and Sugar (2003) proposed B-splines to model the individual curves through mixed effects model where fPC scores are treated as random effects. For sparsely observed data, Yao et al. (2005) proposed the Principal Analysis of Conditional Expectation “PACE” framework. In PACE, fPC scores were estimated by their expectation conditioning on available observations across all trajectories. To estimate fPCs: a system of orthogonal functions, Peng and Paul (2009) proposed a restricted maximum likelihood method based on a Newton–Raphson procedure on the Stiefel manifold. Hall et al. (2006) and Li and Hsing (2010) gave weak and strong uniform convergence rate of the local linear smoother of the mean and covariance, and the rate of derived fPC estimates.
The PACE approach works by efficiently extracting the information on \(\phi _k(t)\) and \(\mu (t)\) even when only a few observations are made on each curve as long as the pooled time points from all curves are sufficiently dense. Nevertheless, PACE is limited by its assumption of independent curves. In reality, observations from different subjects may be correlated. For example, it is expected that expression profiles of genes involved in the same biological processes are correlated; vegetation indices of the same land cover class at neighboring locations are likely to be correlated.
Along with the above approaches, there has been some recent work on correlated functional data. Li et al. (2007) proposed a kernel-based nonparametric method to estimate correlation among functions where observations are sampled at regular temporal grids, and smoothing is performed across different spatial distances. Moreover, it was assumed in their work that the covariance between two observations can be factored as the product of temporal covariance and spatial correlation, which is referred to as separable covariance. Zhou et al. (2010) presented a mixed effect model to estimate correlation structure, which accommodates both separable and nonseparable structures. Mateu (2011) investigated the spatially correlated functional data based on geostatistical and point process contexts and provided a framework for extending multivariate geostatistical approaches in the functional context. Recently, Aston et al. (2015) used a nonparametric approach for estimating the spatial correlation along with providing a test for the separability of the spatial and temporal correlation. Paul and Peng (2011) discussed a nonparametric method similar to PACE to estimate fPCs and proved that the \(L^2\) risk of their estimator achieves optimal nonparametric rate under mild correlation regime when the number of observations per curve is bounded.
Additionally, there has been some parametric approaches toward modeling spatially correlated functional data. For hierarchical functional data which are usually correlated, (Baladandayuthapani et al. 2008) introduced a fully Bayesian method to model both the mean functions and the correlation function in the these data. Additionally, (Staicu et al. 2010) presented fast method to analyze hierarchical functional data when the functions at the deepest level are correlated using multilevel principal components. Later, (Staicu et al. 2012) provided a parametric framework for conducting a study that deals with functional data displaying non-Gaussian characteristics that vary with spatial or temporal location. For parametric models in general, the specific assumptions are very critical to the performance of their respective methods. Gromenko et al. (2012) demonstrated that improved estimation of functional means and Eigen components can be achieved by carefully chosen weighting schemes. Menafoglio and Petris (2016) introduces a very general notion of Kriging for functional data.
In this paper, we develop a new framework which we call SPACE (Spatial PACE) for modeling correlated functional data. In SPACE, we explicitly model the spatial correlation among curves and extend local linear smoothing techniques in PACE to the case of correlated functional data. Our method differs from Li et al. (2007) in that sparsely and irregularly observed data can be modeled, and it is not necessary to assume separable correlation structure. In fact, based on our SPACE framework, we proposed hypothesis tests to examine whether or not correlation structure presented by data is separable.
Specifically, we model the correlation of fPC scores \(s_{ik}\) across curves by anisotropic Matérn family. Matérn correlation model discussed in Abramowitz and Stegun (1966) and Cressie (2015) is widely used in modeling spatial correlation. The anisotropy is introduced by rotating and stretching axes such that equal correlation contour is a tilted ellipse to accommodate anisotropy effects which often arise in geoscience data. In our model, anisotropy Matérn correlation is governed by 4 parameters: \(\alpha ,\delta ,\kappa , \phi \) where \(\alpha \) controls the axis rotation angle and \(\delta \) specifies the amount of axis stretch. SPACE identifies a list of neighborhood structures and applies local linear smoother to estimate a cross-covariance surface for each spatial separation vector. An example of neighborhood structure could be all pairs of locations which are separated by distance of one unit and are positioned from southwest to northeast. In particular, SPACE estimates a cross-covariance surface by smoothing empirical covariances observed at those locations. Next, empirical spatial correlations are estimated based on the eigenvalues of those cross-covariance surfaces. Then, anisotropy Matérn parameters are estimated from the empirical spatial correlations. SPACE directly plugs in the fitted spatial correlation model into curve reconstruction to improve the reconstruction performance relative to PACE where no spatial correlation is modeled, and we show that this procedure is consistent under mild assumptions.
We demonstrate SPACE methodology using simulated functional data as well as on the Harvard Forest vegetation index discussed in Liu et al. (2012). In simulation studies, we first examine the estimation of SPACE model components. Then we perform the hypothesis tests of separability and isotropy effect. We show that curve reconstruction performance is improved using SPACE over PACE. Also, hypothesis tests demonstrate reasonable sizes and powers. Moreover, we construct semi-empirical data by randomly removing observations to achieve sparseness in vegetation index at Harvard Forest. Then it is shown that SPACE restores vegetation index trajectories with fewer errors than PACE.
The rest of the paper is organized as follows. Section 2 describes the spatially correlated functional data model. Section 3 describes the SPACE framework and model selections associated with it. Then we summarize the consistency results of SPACE estimates in Sect. 4 and defer more detailed discussions to the supplementary material. Next, we propose hypothesis tests based on SPACE model in Sect. 5. Section 6 describes simulation studies on model estimations, followed by Sect. 7 which presents curve construction analysis on Harvard Forest data. Conclusions and comments are given in Sect. 8.
2 Correlated functional data model
In this section, we describe how we incorporate spatial correlation into functional data and introduce the Matérn class which we use to model spatial correlation.
2.1 Data generating process
We start by assuming that data are collected across N spatial locations. For location i, a number \(n_i\) of noise-corrupted points are sampled from a random trajectory \(X_i(t)\), denoted by \(Y_i(t_j),\ j = 1,2,\ldots ,n_i\). These observations can be expressed by an additive error model:
Measurement errors \(\{\epsilon _i(t_j)\}_{i=1\ j=1}^{N\ \ \ n_i}\) are assumed to be i.i.d. with variance \(\sigma ^2\) across locations and sampling times. The random function \(X_i(t)\) is the ith realization of an underlying random function X(t) which is assumed to be smooth and square integrable on a bounded and closed time interval \(\mathcal T\). Note that we refer to the argument of function as time without loss of generality. The mean and covariance functions of X(t) are unknown and denoted by \(\mu (t) = E(X(t))\) and \(G(s,t) = \mathrm{Cov}(X(s), X(t))\). By the Karhunen–Loève theorem, under suitable regularity conditions, there exists an eigen decomposition of the covariance kernel G(s, t) such that
where \(\{\phi _k(t)\}_{k=1}^{\infty }\) are orthogonal functions in the \(L^2\) sense which we also call functional principal components (fPCs), and \(\{\lambda _k\}_{k=1}^{\infty }\) are associated nonincreasing eigenvalues. Then, each realization \(X_i(t)\) has the following expansion,
where for given i, \(\xi _{ik}\)’s are uncorrelated fPC scores with variance \(\lambda _k\). Usually, a finite number of eigenfunctions are chosen to achieve reasonable approximation. Then,
In classical functional data model, \(X_i(t)\)’s are independent across i and thus \(cor(\xi _{ik}, \xi _{jk}) = 0\) for any pair of different curves \(\{i,j\}\) and for any given fPC index k. However, in many applications, explicit modeling and estimation of the spatial correlation is desired and can provide insights into subsequent analysis. To build correlation among curves, we assume \(\xi _{ik}\)’s are correlated across i for each k but not across k. One could specify full correlation structure among \(\xi _{ik}\)’s by allowing nonzero covariance between scores of different fPCs, e.g., Cov\((\xi _{ip}, \xi _{jq})\ne 0\). Although the full structure is very flexible, it is subject to the risk of over-fitting and its estimation can become unreliable. Moreover, we believe exposures across curves to the same mode of variation share similarities and tend to co-move more than exposures to different modes of variation do and thus, some parsimony assumptions are reasonable. As a result, we assume that the \(X_i\) can be expressed as a sum of separable processes corresponding to its Eigenfunctions and specify the following structure:
where \(\rho _{ijk}\) measures the correlation between kth fPC scores at curve i and j. Denoting \(\varvec{\xi }_i = (\xi _{i1},\xi _{i2},\ldots ,\xi _{iK})^T\), \(\varvec{\phi }(t) = (\phi _1(t),\phi _2(t),\ldots ,\phi _K(t))^T\) and retaining the first K eigenfunctions as in (2.4), then the covariance between \(X_i(s)\) and \(X_j(t)\) can be expressed as
Note that Cov\((\varvec{\xi }_i\varvec{\xi }_j^T)\) is a diagonal matrix. Hence, columns of \(\varvec{\phi }(t)\) are comprised eigenfunctions of Cov\((X_i(s), X_j(t))\). We also believe that leading fPC scores will be more prone to high correlation than trailing scores, as small variance components tend to exhibit increasingly local features both temporally and spatially. In all subsequent discussions, we assume \(\rho _{ij1}\lambda _1> \rho _{ij2}\lambda _2>\ldots> \rho _{ijK}\lambda _K > 0\). Therefore, in our model specifications, Cov\((\xi _{ip}, \xi _{jq})\) is positive-definite. If we further assume the between-curve correlation \(\rho _{ijk}\) does not depend on k, i.e., \(\rho _{ijk} = \rho _{ij}\), then Cov\((\varvec{\xi }_i\varvec{\xi }_j^T) = \rho _{ij}\mathrm {diag}(\lambda _1,\ldots ,\lambda _K)\). In this case, the covariance can be further simplified as
If the covariance between \(X_i(s)\) and \(X_j(t)\) can be decomposed into a product of spatial and temporal components as in (2.8), we refer to this covariance structure as separable. Separable covariance structure of the noiseless processes assumes that the correlation across curves and across time are independent of each other. One example of this type of processes is the weed growth data studied in Banerjee and Johnson (2006) where curves are weed growth profiles at different locations in the agricultural field. While separable covariance is a convenient assumption which makes estimation easier, it may be unduly restrictive. In order to examine whether this assumption is justifiable, we propose a hypothesis test which is described in Sect. 5. Irrespective of the separability of covariance, spatial correlations among curves are reflected through the correlation structure of fPC scores \(\varvec{\xi }\). For each fPC index k, the associated fPC scores at different locations can be viewed as a spatial random process.
2.2 Matérn class
We choose the Matérn class for modeling spatial correlation. This is widely used as a class of parametric models in geoscience. The Matérn family is attractive due to its flexibility. Specifically, the correlation between two observations at locations separated by distance \(d > 0\) is given by
where \(K_{\nu }(\cdot )\) is the modified Bessel function of the third kind of order \(\nu > 0\) described in Abramowitz and Stegun (1970). This class is governed by two parameters, a range parameter \(\zeta > 0\) which rescales the distance argument and a smoothness parameter \(\nu > 0\) that controls the smoothness of the underlying process.
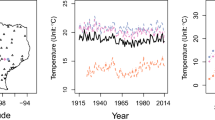
The Matérn class is by itself isotropic which has contours of constant correlation that are circular in two-dimensional applications. However, isotropy is a strong assumption and thus limits the model flexibility in some applications. Liu et al. (2012) showed that fPC scores present directional patterns which might be associated with geographical features. Specifically, the authors look at vegetation index series at Harvard Forest in Massachusetts across a 25 \(\times \) 25 grid over 6 years. Using the same data, we calculate the correlation between fPC scores at locations separated by 45 degree to the northeast and to the northwest, respectively. Figure 1 suggests the anisotropy effect in the second fPC scores. Geometric anisotropy can be easily incorporated into the Matérn class by applying a linear transformation to the spatial coordinates. To this end, two additional parameters are required: an anisotropy angle \(\alpha \) which determines how much the axes rotate clockwise, and an anisotropy ratio \(\delta \) specifying how much one axis is stretched or shrunk relative to the other. Let \((x_1, y_1)\) and \((x_2, y_2)\) be coordinates of two locations and denote the spatial separation vector between these two locations by \(\varvec{\Delta }= (\Delta x, \Delta y)^T = (x_2 - x_1, y_2 - y_1)^T\). The isotropy Matérn correlation is computed typically based on Euclidean distance, and we have \(\rho (d;\zeta ,\nu ) = \rho (\sqrt{\varvec{\Delta }^T\varvec{\Delta }};\zeta ,\nu )\). To introduce anisotropy correlation, a non-Euclidean distance could be applied to variable d through linear transformations on coordinates. Specifically, we form the new spatial separation vector
Spatial correlations of leading fPC scores are computed in directions of northeast-southwest and northwest–southeast separately at Harvard Forest. Consider a two-dimensional coordinates of integers. We calculate the correlation of fPC scores at locations which are positioned along either northwest–southeast or northeast–southwest diagonals, respectively, and are separated by increasing Manhattan distances. Green lines indicate the correlations along the northwest–southeast diagonal and red lines indicate the correlations along the northeast–southwest diagonal. The first two leading fPCs are illustrated. (Color figure online)
where we write the rotation and rescaling matrix by \(\mathbf{R}\) and \(\mathbf{S}\) respectively. Define the non-Euclidean distance function as \(d^*(\varvec{\Delta },\alpha ,\delta ) = \sqrt{\varvec{\Delta }^{*T}\varvec{\Delta }^*} = \sqrt{\varvec{\Delta }^T\mathbf{R}^T\mathbf{S}^2\mathbf{R}\varvec{\Delta }}\). Hence, the anisotropy correlation is computed as
Note \(\rho ^*(\varvec{\Delta }) = \rho ^*(-\varvec{\Delta })\). Let \(\varvec{\Delta }_{ij}\) be the spatial separation vector of locations between-curve i and j. Then we model the covariance structure of fPC scores described in (2.5) as
Note that the above parametrization is not identifiable. First, \(\alpha \) and \(\alpha \) + \(\pi \) always give the same correlation. Second, the pair of any \(\alpha \) and \(\delta \) gives the exact same correlation with the combination of \(\alpha + \pi /2\) and \(1/{\delta }\). We remove the nonuniqueness of model by adding additional constraints on the ranges of parameters.
3 SPACE methodology
SPACE methodology extends the PACE methodology introduced by Yao et al. (2005) and the methodology discussed in Li et al. (2007). Among the components of SPACE, the mean function \(\mu (t)\) and the measurement error variance \(\sigma ^2\) in (2.1) are estimated by the same method used in Yao et al. (2005). In this section, we introduce the estimation of the other key components of SPACE: the cross-covariance surface Cov\((X_i(s), X_j(t))\) for any location pair (i, j) in (2.6) and the anisotropy Matérn model among fPC scores in (2.12). We will also describe methods of curve reconstruction and model selection.
3.1 Cross-covariance surface
Let us define \(G_{ij}(s,t) = \mathrm{Cov}(X_i(s), X_j(t))\) as the cross- covariance surface between location i and j. Let \(\hat{\mu }(t)\) be an estimated mean function, and then let \(D_{ij}(t_{ik},t_{jl}) = (Y_i(t_{ik}) - \hat{\mu }(t_{ik}))(Y_j(t_{jl}) - \hat{\mu }(t_{jl}))\) be the raw cross-covariance based on observations from curve i at time \(t_{ik}\) and curve j at time \(t_{jl}\). We estimate \(G_{ij}(s,t)\) by smoothing \(D_{ij}(t_{ik},t_{jl})\) using a local linear smoother. Gromenko et al. (2012) have demonstrated that improved estimation of both mean and covariance can be improved by accounting for spatial correlation between curves. Here we have chosen simpler estimates for the sake of presentation, but the tests designed below can be combined with any estimate of the mean and eigenfunctions of our sample.
In this work, we assume the second-order spatial stationarity of the fPC score process. Define \(N(\varvec{\Delta }) = \{(i,j),\ s.t.\ \varvec{\Delta }_{ij} = (\Delta _x, \Delta _y)\ {\text {or}}\ \varvec{\Delta }_{ij} = (-\Delta _x, -\Delta _y)\}\) to be the collection of location pairs that share the same spatial correlation structure. Then, all location pairs that belong to \(N(\varvec{\Delta })\) are associated with the same unique covariance surface which we write as \(G_{{\varvec{\varDelta }}}(s,t)\). As a result, all raw covariances constructed based on locations in \(N(\varvec{\Delta })\) can be pooled together to estimate \(G_{{\varvec{\Delta }}}(s,t)\). In some cases, the number of elements in \(N({\varvec{\Delta }})\) is very limited. For example, if observations are collected from irregular and sparse locations, it is relatively rare for two pair of locations to have exactly the same spatial separation vector. More location pairs can be included by working with a sufficiently small neighborhood around a given \(\varvec{\Delta }\). Define \(N_B(\varvec{\Delta }) = \{(i,j),\ s.t. \varvec{\Delta }_{ij}\in B\}\), where B as an appropriately specified neighborhood “ball” centering around \(\varvec{\Delta }\). The estimate of cross-covariance surface can be derived by replacing \(N(\varvec{\Delta })\) with \(N_B(\varvec{\Delta })\) in (3.2). Other estimates of cross-covariance could be obtained based on smoothing over distances. This would require the selection of a window width; since our examples all involve regularly spaced data, we do not pursue this here. In addition, we note
where \(\text{ I }(i = j, s = t)\) is 1 if \(i = j\) and \(s = t\), and 0 otherwise. If \(i = j\), the problem reduces to the estimation of covariance surface, and we apply the same treatment described in Yao et al. (2005) to deal with the extra \(\sigma ^2\) on the diagonal. For \(i\ne j\) and a given spatial separation vector \(\varvec{\Delta }\), the local linear smoother of the cross-covariance surface \(G_{{\varvec{\Delta }}}(s,t)\) is derived by minimizing
with respect to \(\beta _0,\beta _1\), and \(\beta _2\). \(\kappa _2\) is the two-dimensional Gaussian kernel. Let \(\hat{\beta }_0\), \(\hat{\beta }_1\), and \(\hat{\beta }_2\) be minimizers of (3.2). Then \(\widehat{G}_{{\varvec{\Delta }}}(s,t) = \hat{\beta }_0\). For computation, we evaluate one-dimensional functions over equally spaced time points \({\mathbf{t}^{\mathrm{eval}}} = (t_1^{\mathrm{eval}},\cdots ,t_M^{\mathrm{eval}})^T\) with step size h between two consecutive points. We evaluate \(\widehat{G}_{{\varvec{\Delta }}}(s,t)\) over all possible two-dimensional grid points constructed from \({\mathbf{t}^{\mathrm{eval}}}\), denoted by \(\mathbf{\mathbf{t}^{\mathrm{eval}}}\times {\mathbf{t}^{\mathrm{eval}}}\). Let \(\widehat{G}_{{\varvec{\Delta }}}({\mathbf{t}^{\mathrm{eval}}}\times {\mathbf{t}^{\mathrm{eval}}})\) be the evaluation matrix across all grid points. The estimates of eigenfunction \(\phi _k(t)\) and \(\lambda _k\) are derived as the eigenvectors and eigenvalues of \(\widehat{G}_{{\varvec{\Delta }}}({\mathbf{t}^{\mathrm{eval}}}\times {\mathbf{t}^{\mathrm{eval}}})\) adjusted for the step size h.
3.2 Anisotropy Matérn model
Now we focus on estimating the parameters of the Matérn model. We will first estimate the empirical correlation \(\rho _k(\varvec{\Delta })\) for the cross-covariance surfaces and estimate the parameters of the Matérn model by fitting them to the empirical correlations. Equation (2.12) specifies the spatial covariance among fPC scores. Let \(\lambda _k(\varvec{\Delta })\) be the kth eigenvalue of \(G_{{\varvec{\Delta }}}(s,t)\). For covariance surface, we use \(\lambda _k((0,0))\) and \(\lambda _k\) interchangeably. Then we have
If we further assume \(\rho ^*_1(\varvec{\Delta })\lambda _1> \rho ^*_2(\varvec{\Delta })\lambda _2>\ldots>\rho ^*_K(\varvec{\Delta })\lambda _K > 0\), then the sequence \(\left\{ \rho ^*_k(\varvec{\Delta })\lambda _k\right\} _{k=1}^K\) are eigenvalues of \(G_{{\varvec{\Delta }}}(s,t)\) ordered from the largest to the smallest. Note that \(\rho ^*_k(\varvec{\Delta }) = 1\) if \(\varvec{\Delta }= (0,0)\) for all k. Thus for all \(\varvec{\Delta }> 0\), \(\rho ^*_k(\varvec{\Delta })\) can be estimated as the ratio of kth eigenvalues of \(G_{{\varvec{\Delta }}}(s,t)\) and \(G_{(0,0)}(s,t)\), which can be written as
where \(\hat{\lambda }_k(\varvec{\Delta })\) is the kth eigenvalue of \(\widehat{G}_{{\varvec{\Delta }}}\) and \(\hat{\lambda }_k\) is the kth eigenvalue of \(\widehat{G}_{(0,0)}\). Suppose empirical correlations \(\left\{ \hat{\rho }^*_k(\varvec{\Delta }_i)\right\} _{i=1}^{m}\) are obtained for \(\left\{ \varvec{\Delta }_i\right\} _{i=1}^m = \left\{ \varvec{\Delta }_1,\ldots ,\varvec{\Delta }_m\right\} \). Then
are used to fit (2.11) and to estimate parameters \(\alpha ,\delta ,\zeta ,\nu \). If assuming separable covariance structure, empirical correlations could be pooled across k to estimate parameters of the anisotropy Matérn model. If \(\left\{ B(\varvec{\Delta }_i,\varvec{\delta }_i)\right\} _{i=1}^m\) are used, then we select one representative vector from each \(B(\varvec{\Delta }_i,\varvec{\delta }_i)\) as input. A sensible choice of representative vectors is just \(\left\{ \varvec{\Delta }_i\right\} _{i=1}^m\), the center of \(\left\{ B(\varvec{\Delta }_i,\varvec{\delta }_i)\right\} _{i=1}^m\). When fitting (2.11), the sum of squared difference between empirical and fitted correlations over all \(\varvec{\Delta }_i\)’s is minimized through numerical optimization. We adopt BFGS method in implementation. More details about the quasi-Newton method can be found in Avriel (2003).
3.3 Curve reconstruction
Reconstructing trajectories are the important applications of the SPACE model. Curve reconstructions based on SPACE model also provide an alternative perspective of “gap-filling” the missing data for geoscience applications as well. Equation (2.4) specifies the underlying formula used to reconstruct the trajectory \(X_i(t)\) for each i. \(\{\hat{\varvec{\phi }}_k(t)\}_{k=1}^K\) and \(\hat{\mu }(t)\) are obtained from the estimation of the covariance surface in a manner analogous to PACE, as described in the previous section. We have found empirically that although the cross-covariance surfaces share the same eigenfunctions, these do not perform as well as those obtained from the covariance only. The only missing element now is fPC scores \(\{\xi _{ik}\}_{i=1,k=1}^{N,K}\). The best linear unbiased predictors (BLUP) (Ruppert et al. 2003) of \(\xi _{ik}\) are given by
To describe the closed-form solution to Eq. (3.6) and to facilitate subsequent discussions, we introduce the following notations. Write \(\mathbf{Y}_i = (Y_i(t_{i1}),\ldots ,Y_i(t_{in_i}))^T\), \(\widetilde{\mathbf{Y}} = (\mathbf{Y}_1,\ldots ,\mathbf{Y}_N)^T\), \({\varvec{\mu }}_i = (\mu (t_{i1}),\ldots ,\mu (t_{in_i}))^T\), \(\widetilde{\varvec{\mu }} = ({\varvec{\mu }}_1,\ldots ,{\varvec{\mu }}_N)^T\), \({\varvec{\xi }}_i = ({\xi _{i1}},\ldots ,{\xi _{iK}})^T\), \(\widetilde{\varvec{\xi }} = ({\varvec{\xi }}_1,\ldots ,{\varvec{\xi }}_N)^T\), \({\varvec{\Lambda }} = diag(\lambda _1,\ldots ,\lambda _K)\), \(\rho ^*_{ijk} = \rho ^*_k(\varvec{\Delta }_{ij})\), \({\varvec{\rho }}_{ik} = (\rho ^*_{i1k},\ldots ,\rho ^*_{iNk})^T\), \({\varvec{\rho }}_k = ({\varvec{\rho }}_{1k}^T,\ldots ,{\varvec{\rho }}_{Nk}^T)^T\), \({\varvec{\rho }}_{ij}=diag(\rho ^*_{ij1},\ldots ,\rho ^*_{ijK})\), \(\widetilde{\varvec{\rho }} = \left[ {\varvec{\rho }}_{ij}\right] \), where  represents a matrix with ijth entry equal to
represents a matrix with ijth entry equal to  , and \(\varvec{\phi }_{ik} = (\phi _k(t_{i1}),\ldots ,\phi _k(t_{in_i}))^T\), \({\varvec{\phi }}_i = (\varvec{\phi }_{i1},\ldots ,\varvec{\phi }_{iK})\), \(\widetilde{\varvec{\phi }} = diag({\varvec{\phi }}_1,\ldots ,{\varvec{\phi }}_N)\). Note diagonalization and transpose are performed before substitution in all above notations. If assuming separable covariance, we write \({\varvec{\rho }} = \left[ \rho ^*_{ij}\right] \). In addition, define \(\varvec{\Sigma }(A,B)\) as the covariance matrix between A and B, and \(\varvec{\Sigma }(A)\) as the variance matrix of A. Then
, and \(\varvec{\phi }_{ik} = (\phi _k(t_{i1}),\ldots ,\phi _k(t_{in_i}))^T\), \({\varvec{\phi }}_i = (\varvec{\phi }_{i1},\ldots ,\varvec{\phi }_{iK})\), \(\widetilde{\varvec{\phi }} = diag({\varvec{\phi }}_1,\ldots ,{\varvec{\phi }}_N)\). Note diagonalization and transpose are performed before substitution in all above notations. If assuming separable covariance, we write \({\varvec{\rho }} = \left[ \rho ^*_{ij}\right] \). In addition, define \(\varvec{\Sigma }(A,B)\) as the covariance matrix between A and B, and \(\varvec{\Sigma }(A)\) as the variance matrix of A. Then

where \(\otimes \) represents Kronecker product. With Gaussian assumptions, we have
where the last line follows the Woodbury matrix identity. For cases where \(\sum _{i=1}^N n_i > NK\), the transformation of last line suggests a way to reduce the size of matrix to be inverted. The separability assumption simplifies not only the model itself but the calculation of matrix inverse as well, noting that \(\varvec{\Sigma }(\widetilde{\varvec{\xi }},\widetilde{\varvec{\xi }})^{-1} = ({\varvec{\rho }}\otimes {\varvec{\Lambda }})^{-1} = {\varvec{\rho }}^{-1}\otimes {\varvec{\Lambda }}^{-1}\). By substituting all elements in (3.8) with corresponding estimates, the estimate of \(\widetilde{\varvec{\xi }}\) is derived as

The reconstructed trajectory is then given by
While we start with binned estimates of correlation, the use of a parametric spatial correlation kernel allows us to regularize these estimates, particularly at larger distances and when the data are not regularly spaced. It also allows us to obtain an orientation from which to test isotropy.
3.4 Model selection
Rice and Silverman (1991) proposed a leave-one-curve-out cross-validation method for data which are curves. Hurvich et al. (1998) introduced a methodology for choosing smoothing parameter for any linear smoother based on an improved version of Akaike information criterion (AIC). Yao et al. (2005) pointed out that adaptation to estimated correlations when estimating the mean function with dependent data does not lead to improvements and subjective choice are often adequate.
In local linear smoothing of both cross-covariance surface and mean curve, we use the default cross-validation method employed in the sm package (Bowman and Azzalini 2013) of (R Development Core Team 2010), in particular, that method groups observations into bins and perform leave-one-bin-out cross-validation (LOBO). We also examine other alternatives which include a) leave-one-curve-out cross-validation of Rice and Silverman (1991) (LOCO), b) J-fold leave-curves-out cross-validation (\(\mathrm {LCO_J}\)) and c) the improved AIC method of Hurvich et al. (1998). In all cross-validation methods, the goodness of smoothing is assessed by squared prediction error. In simulations not reported here, LOBO method demonstrates the most consistent performance in terms of eigenvalue estimation. Sparseness and noise of observations will inflate estimated eigenvalues up whereas smoothing tends to shrink estimates down. LOBO method achieves better balance between those two competing forces. We leave a more detailed investigation of this phenomenon to future work.
To determine the number of eigenfunctions K which sufficiently approximate the infinite dimensional process, we rely on \(\mathrm {LCO_J}\) of curve reconstruction error which is denoted by \(\mathrm {ErrCV_J}\) for convenience. Specifically, all curves are partitioned into J complementary groups. Curves in each group serve as the testing sample. Then we select the training sample as curves that are certain distance away from testing curves to reduce the spatial correlation between the training and testing samples. For each fold, testing curves are reconstructed based on parameters estimated by corresponding training curves. Denote reconstructed curves of the jth fold by \(\widehat{\widetilde{\mathbf{X}}}_{-j}\). Then, we compute \(\mathrm {ErrCV_J}\) as follows:
Zhou et al. (2010) pointed out that cross-validation scores may keep decreasing as model complexity increases, which is also observed in our simulation studies. This was explained as an effect of using an approximate model particularly for covariance; we hypothesize that residual spatial dependence will compensate for additional model complexity without actually improving predictive error. The 5-fold cross-validation score, \(\mathrm {ErrCV_5}\), as a function of K is illustrated in Fig. 2. A quick drop is observed followed by a much slower decrease. Instead of finding the minimum \(\mathrm {ErrCV_5}\), we visually select a suitable K as the kink of \(\mathrm {ErrCV_5}\) profile. In Fig. 2, the largest drop of CV scores takes place at the correct value \(K = 2\) for 100 % times out of the 200 replicated datasets.
Leave-curves-out cross-validation. Simulated curves are constructed as the linear combination of eigenfunctions over time interval [0,1]. We use two eigenfunctions \(\phi _1(t)\equiv 1\) and \(\phi _{2}(t)=\sin (\omega t)\). Two separable spatial correlation structures are generated over a one-dimensional horizontal array of 100 equally spaced locations. Eigenvalues are \(\lambda _1 = \exp (-1)\times 10 = 3.679\) and \(\lambda _2 = \exp (-2)\times 10 = 1.353\). Matérn parameters are \(\alpha = 0, \delta = 1, \zeta = 8, \nu = 0.5\) for high correlation scenario and \(\alpha = 0, \delta = 1, \zeta = 1, \nu = 0.5\) for low correlation scenario. Corresponding correlations at distance 1 are 0.8825 and 0.3679. Noise standard deviation is \(\sigma = 1\). When estimating model parameters, we use two spatial separation vectors \({\varvec{\Delta }}_1 = (1,0)\) and \({\varvec{\Delta }}_2 = (2,0)\). \(\mathrm {ErrCV_5}\) is computed over 200 simulated datasets with number of eigenfunctions \(K = 1,2,3,4\). Median \(\mathrm {ErrCV_5}\) is represented by solid line and 5 and 95 % percentiles are plotted in dashed lines
When fitting anisotropy Matérn model, \(\left\{ \varvec{\Delta }_i\right\} _{i=1}^m\), the set of neighborhood structures needs to be determined. Consider the example of one-dimensional and equally spaced locations where the ith location has coordinates (i, 0). Larger spatial separation vector corresponds to fewer pairs of locations in \(N(\varvec{\Delta })\). Thus we often start with the most immediate neighborhood and choose \(\left\{ \varvec{\Delta }_i\right\} _{i=1}^m = \left\{ (1,0),(2,0),\ldots ,(m,0)\right\} \) with m to be decided. In simulation studies not reported here, we found that \(\mathrm {ErrCV_J}\) is not sensitive with respect to m, which is especially true when spatial correlation is low. Instead of selecting an “optimal” m, we use a range of different m’s and take the trimmed average of estimates derived across m. The maximum m can be selected so that the spatial correlation is too low to have meaningful impact based on prior knowledge. In simulation studies of Sect. 6, all estimations are made based on 20 % (each side) trimmed mean across a list of \(\left\{ \varvec{\Delta }_i\right\} _{i=1}^m\)’s.
4 Asymptotic properties of estimates
Assuming no spatial correlation, Yao et al. (2005) demonstrated the consistency of estimated fPC scores \(\hat{\xi }_{ik}\), reconstructed curves \(\widehat{X}_i(t)\) and other model components. In Paul and Peng (2011), authors proposed a nonparametric and kernel-based estimator of functional eigenstructure similar to Yao et al. (2005). It was shown in their work that if the correlation between sample curves is “weak” in an appropriate sense, their estimator converges at the same rate in correlated and i.i.d. cases. The uniform convergence of the local linear smoothers of mean and covariance functions on bounded intervals plays a central role in the derivation of asymptotic results in Yao et al. (2005). We show that the same rate of convergence stated in their work can be achieved with the presence of sufficiently mild spatial correlation. For smoother of mean function, we require the following:
- (D1):
-
$$\begin{aligned}&\dfrac{1}{N^2}\sum _{1\le i_1\ne i_2\le N}|\rho ^*_k\left( \varvec{\Delta }_{i_1i_2}\right) |\rightarrow 0\ \mathrm{as}\ N\rightarrow \infty ,\ \mathrm{for}\\&\quad k = 1,2,\cdots ,K. \end{aligned}$$
Condition (D1) indicates that for every eigenfunction index k, the spatial correlation must decay sufficiently fast as distance increases so that the average spatial correlation across all location pairs becomes negligible as the number of curves goes to infinity.
In the case of cross-covariance smoother, we consider two pairs of locations \((i_1,j_1)\) and \((i_2,j_2)\), both of which have the same spatial separation vector \(\varvec{\Delta }_0\), i.e., \((i_1,j_1)\in \mathcal{N}(\varvec{\Delta }_0)\) and \((i_2,j_2)\in \mathcal{N}(\varvec{\Delta }_0)\). Denote the spatial separation vectors of pairs \((i_1,i_2)\), \((i_1,j_2)\) and \((j_1,i_2)\) by \(\varvec{\Delta }_1\), \(\varvec{\Delta }_2\) and \(\varvec{\Delta }_3\), respectively. The following sufficient condition is required for cross-covariance smoother:
- (D2):
-

Note that locations \(i_1, i_2, j_1\), and \(j_2\) form a parallelogram in two-dimensional space. \(\varvec{\Delta }_0\) and \(\varvec{\Delta }_1\) represent vectors of two edges and \(\varvec{\Delta }_2\) and \(\varvec{\Delta }_3\) represent vectors of two diagonals. Two edge vectors determine the shape of parallelogram and diagonals can be derived from edges. In (D2), \(\varvec{\Delta }_0\) is fixed in the summation and the only degree of freedom is \(\varvec{\Delta }_1\) which essentially plays the same role as \(\varvec{\Delta }_{ij}\) in (D1). Therefore, a specific spatial correlation structure is likely to satisfy both (D1) and (D2) at the same time. Consider a spatial AR(1) correlation structure on a line as example where locations \(i_1, i_2, j_1 \), and \(j_2\) are grid points on a line and \(\rho ^*_k(\varvec{\Delta }_{ij}) = \rho ^{|i-j|}\ \forall k\). Then the parallelogram degenerates and the summation in (D2) reduces to

It is easy to show that AR(1) correlation satisfies both (D1) and (D2).
Based on the framework of asymptotic properties in Yao et al. (2005) and above sufficient conditions, we extend their results to correlated case and state the consistency of \({\hat{\mu }}(t)\) and \({\widehat{G}}_{\tiny {\varvec{\Delta }}}(s,t)\) in the following theorem.
Theorem 1* Under (A1.1)–(A4) and (B1.1)–(B2.2b) in Yao et al. (2005) with \(\nu =0,l=2\) in (B2.2a), \(\nu =\mathrm{(}0,0\mathrm{)},l=2\) in (B2.2b), and (D1)–(D2),
and
The proof of Theorem 1* and more discussions can be found in the supplementary material. In appendices, we first review the main theorems and underlying assumptions discussed in Yao et al. (2005). Then we provide the proof of Theorem 1*. Following the same line of arguments as in Yao et al. (2005), it is straightforward to derive the counterparts of remaining results in Yao et al. (2005) accordingly based on Theorem 1*.
5 Separability and isotropy tests
Separability of spatial covariance is a convenient assumption under which Cov\(({\varvec{\xi }}_i, {\varvec{\xi }}_j)\) is simply a rescaled identity matrix and thus a parsimonious model could be fitted. However, we would like to design a hypothesis test to examine if this assumption is valid and justified by the data. Spatial correlation could depend not only on distance but angle as well. Whether correlation is isotropic or not may be an interesting question for researchers and is informative for subsequent analysis. In this section, we propose two hypothesis tests to address these issues based on SPACE model.
5.1 Separability test
Recall the correlation matrix of the kth fPC scores among curves is denoted by \({\varvec{\rho }}_k\). We model \({\varvec{\rho }}_k\) through anisotropy Matérn model through parameters \(\alpha _k,\delta _k,\zeta _k\), and \(\nu _k\). Suppose we use K eigenfunction to approximate the underlying process. Then we partition the set \(\{1,2,\ldots ,K\}\) into A mutually exclusive and collectively exhaustive groups \(\mathbf{K} = \{\mathcal{K}_1,\ldots , \mathcal{K}_A\}\). Denote the number of k’s in group \(\mathcal{K}_i\) by \(|\mathcal{K}_i|\). A generic hypothesis can be formulated in which parameters across k’s are assumed to be the same within each group but can be different between groups. Consider a simple example where we believe correlation structures are the same among the first three fPC scores versus the hypothesis that correlations are all different. Then the partition associated with the null hypothesis is \(\mathbf{K}^0:\{\mathcal{K}_1=\{k_1,k_2,k_3\},\mathcal{K}_2=\{k_4\},\ldots ,\mathcal{K}_{K-2}=\{K\}\}\). The partition for alternative hypothesis is \(\mathbf{K}^1:\{\mathcal{K}_1=\{k_1\},\mathcal{K}_2=\{k_2\},\ldots ,\mathcal{K}_{K}=\{K\}\}\). Both the null and alternative can take various forms of partitions. However, for illustration purpose, we consider the following test where no constraints are imposed in the alternative:
The key step of the test is to construct hypothesized null curves from observed data. If sample curves are independent, hypothesized null curves can be constructed through bootstrapping fPC scores, see Liu et al. (2012) and Li and Chiou (2011). With correlated curves, reshuffling of the fPC scores is not appropriate as correlation structure would break down. However, correlated fPC scores can be transformed to uncorrelated z-scores based on the covariances estimated from anisotropy Matérn model. Then z-scores are reshuffled by curve-wise bootstrapping. Next, reshuffled z-scores are transformed back to the original fPC score space based on the covariances estimated under the null hypothesis. Then hypothesized null curves are constructed as the linear combination of eigenfunctions weighted by resampled fPC scores. Let \({\varvec{\theta }}_k\triangleq \{\alpha _k,\delta _k,\zeta _k,\nu _k\}\) be the set of anisotropy Matérn parameters. The detailed procedure is described as follows.
- Step 1 :
-
Estimate model parameters assuming they are arbitrary. Denote the estimates by \({\hat{\sigma }}^2,\widehat{\varvec{\phi }}(t),\widehat{\varvec{\lambda }}\), and \(\widehat{\varvec{\theta }}_k\). Then compute the observed test statistics \(\mathcal{S}\) using these estimates.
- Step 2 :
-
Estimate fPC scores using \({\hat{\sigma }}^2,\widehat{\varvec{\phi }}(t),\widehat{\varvec{\lambda }}\), and \(\widehat{\varvec{\theta }}_k\) assuming nonseparable case in (3.11). Note \(\widehat{\widetilde{\varvec{\xi }}}\) in (3.11) is arranged by curve index i. We rearrange it by fPC index k and let \(\widehat{\varvec{\xi }}_k\) be the vector of estimated kth fPC scores across all locations.
- Step 3 :
-
Estimate \({\varvec{\theta }}_k\) assuming \(\mathbf{H}_0\). Specifically, for each r, fit anisotropy Matérn model with pooled inputs \(\{\varvec{\Delta }_l,\hat{\rho }_k^*({\varvec{\Delta }}_l),\forall k\in \mathcal{K}_r\}_{l=1}^L\). Let \(\widehat{\varvec{\theta }}_k^{\tiny {0}}\) be the pooled estimates. Note \(\widehat{\varvec{\theta }}_{k_1}^{\tiny {0}} = \widehat{\varvec{\theta }}_{k_2}^{\tiny {0}}\) if \(k_1,k_2\) are both in \(\mathcal{K}_r\).
- Step 4 :
-
For each k whose associated \(\mathcal{K}_r\) has more than one indices, let \(\widehat{{\varvec{\rho }}}_k\) be the spatial correlation matrix constructed using \(\widehat{\varvec{\theta }}_k^{\tiny {0}}\). Suppose \(\widehat{{\varvec{\rho }}}_k\) has eigen decomposition \(\mathbf{VCV^{\tiny {T}}}\). Then \(\widehat{\mathrm{Cov}}({\varvec{\xi }}_k) = \hat{\lambda }_k\mathbf{VCV^{\tiny {T}}}\). Define transformation matrix \({\mathbf{t}} = ({\hat{\lambda }}_k\mathbf{C})^{-1/2}{} \mathbf{V}^{\tiny {T}}\) and the transformed scores \({\widehat{\mathbf{z}}}_k = {\mathbf{t}}\widehat{\varvec{\xi }}_k\). Then \(\widehat{\mathrm{Cov}}({\widehat{\mathbf{z}}}_k) = \mathbf{I}_{N\times N}\).
- Step 5 :
-
Resample \({\widehat{\mathbf{z}}}_k\)’s through curve-wise bootstrapping. Specifically, randomly sample the curve indices \(\{1,\cdots ,N\}\) with replacement and let P(i) be the permuted index for curve i. Then the bth bootstrapped score for curve i is obtained as \({\hat{z}}^{\tiny {b}}_{ik} = {\hat{z}}_{{\tiny {P(i)}}k}\) for all k’s such that \(|\mathcal{K}_r| > 1\).
- Step 6 :
-
Transform \({\widehat{\mathbf{z}}}^{\tiny {b}}_k\)’s back to the space of fPC scores. Define resampled fPC scores \({\widehat{\varvec{\xi }}}^{\tiny {b}}_k = {\mathbf{t}}^{-1}{\widehat{\mathbf{z}}}^{\tiny {b}}_k\).
- Step 7 :
-
Generate the bth set of Gaussian random noises \(\{\epsilon ^{\tiny {b}}(t_{ij})\}_{i=1,j=1}^{N,n_i}\) based on the noise standard deviation \({\hat{\sigma }}^2\) estimated in step 1.
- Step 8 :
-
Construct resampled observations as below
$$\begin{aligned}&Y_{i}^{\tiny {b}}(t_{ij}) = \hat{\mu }(t_{ij})+ \sum _{|\mathcal{K}_r| > 1}\sum _{k\in \mathcal{K}_r}\hat{\xi }^{\tiny {b}}_{ik}\hat{\phi }_k(t_{ij}) \nonumber \\&\qquad \qquad \qquad + \sum _{|\mathcal{K}_r| = 1}\sum _{k\in \mathcal{K}_r}\hat{\xi }_{ik}\hat{\phi }_k(t_{ij}) + \epsilon ^{\tiny {b}}(t_{ij}) \end{aligned}$$(5.3) - Step 9 :
-
Given \(\{Y_{i}^{\tiny {b}}(t_{ij})\}_{i=1,j=1}^{N,n_i}\), estimate anisotropy Matérn parameters. Denote the estimates by \(\widehat{\varvec{\theta }}^{\tiny {b}}_k\). Then compute test statistics \(\mathcal{S}^{\tiny {b}}\).
- Step 10 :
-
Repeat steps 5–9 for B times and obtain \(\{\mathcal{S}^{\tiny {b}}\}_{b=1}^{B}\) which form the empirical null distribution of the test statistics. Then make decision by comparing the null distribution with the observed test statistics \(\mathcal{S}\).
Note that an alternative and theoretically more precise way of doing step 4 is to use covariances constructed based on \(\widehat{\varvec{\theta }}_k\) as opposed to \(\widehat{\varvec{\theta }}_k^{\tiny {0}}\). The alternative way is assumed to remove spatial correlation more thoroughly but simulation results not reported suggest that it introduces more volatility into z-scores that offsets the benefit of lower residual spatial correlation in them. For testing the \(\mathbf{H}_0\) above, we transform \(\widehat{\varvec{\theta }}_k\) and \(\widehat{\varvec{\theta }}^{\tiny {b}}_k\) from parameter space to correlation space. Let \(\bar{\rho }^*_r({\varvec{\Delta }}^{\mathrm{eval}}; \widehat{\varvec{\theta }}_k) = \sum _{k\in \mathcal{K}_r}\rho ^*({\varvec{\Delta }}^{\mathrm{eval}}; \widehat{\varvec{\theta }}_k)/{|\mathcal{K}_r|}\) be the average correlation computed at separation vector \({\varvec{\Delta }}^{\mathrm{eval}}\) across k’s in \(\mathcal{K}_r\). Then define test statistics as
Our experience from a preliminary simulation study with different parameter settings has been that statistics in correlation space have greater power than those in parameter space, although these are similar in our studies below. In our implementations, we choose \({\varvec{\Delta }}^{\mathrm{eval}}\) to be the most immediate neighborhood in Euclidean distance. This correlation is computed from the Matern covariance, and we have chosen this distance as being likely to exhibit the largest distinction between Eigen components.
5.2 Isotropy test
Isotropy test is essentially the same as the separability test. To make the test more general, we assume a prior that correlation structures are the same across k’s within \(\mathcal{K}_r\) for \(r = 1,2,\ldots ,A\). Note that within each \(\mathcal{K}_r\), \(\alpha _k\)’s and \(\delta _k\)’s must take equal values across k’s. Let \(\mathbf{K}_a = \{\mathcal{K}_{r_1},\ldots ,\mathcal{K}_{r_a}\}\) be the set whose \(\alpha _k\)’s are all assumed to be zeros. Then let \(\mathbf{K}_{a}^c\) be the complement set of \(\mathbf{K}_{a}\) where \(1 \le a\le A\). To test the following hypothesis
we propose a similar procedure which slightly differs from the separability test in step 1, step 3, step 7, and step 8. Specifically, we replace those steps in separability tests with the following.
- Step 1* :
-
Estimate model parameters under the prior. In particular, \(\widehat{\varvec{\theta }}_k\) is obtained by pooling empirical correlations from all k’s within \(\mathcal{K}_r\). Then compute \(\mathcal{S}\) based on these estimates.
- Step 3* :
-
Under the null hypothesis, we estimate anisotropy Matérn parameters. For any \(k\in \mathcal{K}_r\) that belongs to \(\mathbf{K}_a\), \(\hat{\alpha }_k^{\tiny {0}}\) and \(\hat{\delta }_k^{\tiny {0}}\) are fixed at zero and one, respectively, whereas \({\hat{\zeta }}_k^{\tiny {0}}\) and \({\hat{\nu }}_k^{\tiny {0}}\) are estimated from \(\{\varvec{\Delta }_l,\hat{\rho }_k^*({\varvec{\Delta }}_l),\forall k\in \mathcal{K}_r\}_{l=1}^L\). Then \(\widehat{\varvec{\theta }}_k^{\tiny {0}} = (0,1,{\hat{\zeta }}_k^{\tiny {0}},{\hat{\nu }}_k^{\tiny {0}})\) for any \(k\in \mathcal{K}_r\) that belongs to \(\mathbf{K}_a\). For any \(k\in \mathcal{K}_r\) that belongs to \(\mathbf{K}_a^c\), no extra estimation is needed. We keep fPC scores estimated in step 2 for those k’s and perform no transformation on them.
- Step 8* :
-
Construct resampled curves as below
$$\begin{aligned}&Y_{i}^{\tiny {b}}(t_{ij}) = \hat{\mu }(t_{ij}) + \sum _{\mathcal{K}_r\in \mathbf{K}_a}\sum _{k\in \mathcal{K}_r}\hat{\xi }^{\tiny {b}}_{ik}\hat{\phi }_k(t_{ij}) \nonumber \\&\quad + \sum _{\mathcal{K}_r\in \mathbf{K}^c_a}\sum _{k\in \mathcal{K}_r}\hat{\xi }_{ik}\hat{\phi }_k(t_{ij}) + \epsilon ^{\tiny {b}}(t_{ij}). \end{aligned}$$(5.8) - Step 9* :
-
Given \(\{Y_{i}^{\tiny {b}}(t_{ij})\}_{i=1,j=1}^{N,n_i}\), estimate anisotropy Matérn parameters.
Accordingly, the test statistics \(\mathcal{S}\) could be as simple as the sum of absolute \(\hat{\alpha }\)’s,
where \({\hat{\alpha }}_r\) represents the common estimate for all k’s in set \(\mathcal{K}_r\). Testing procedures described above can be easily extended to the case where both separability and isotropy effect are of interest.
6 Simulations
In this section, we present simulation studies of the SPACE model. We examine the estimation of anisotropy Matérn parameters and curve reconstructions in Subsect. 6.1. Simulation results of hypothesis tests are shown in Subsect. 6.2. For both one-dimensional and two-dimensional locations, we consider grid points with integer coordinates. Specifically, two-dimensional coordinates take the form of \(\{(i,j), 1\le i,j\le E\}\), where E is referred to as the edge length. Similarly, one-dimensional locations are represented by \(\{(0,1),(0,2),\ldots ,(0,N)\}\) where N is the total number of locations. For simplicity, we examine a subset of anisotropy Matérn family by fixing \(\nu \) at 0.5, which corresponds to the spatial autoregressive model of order one. Below we have treated \(\nu \) as fixed as corresponding to an interpretable structure. This follows the recommendation in Diggle et al. (2003); in practice, the determination of \(\nu \) from real data can be difficult. When constructing simulated curves, we employ two eigenfunctions which are \(\phi _1(t) = 1\) and \(\phi _2(t) = \sin (2\pi t)\), with eigenvalues \(\lambda _1 = \exp (-1)\times 10 \approx 3.68\) and \(\lambda _2 = \exp (-2)\times 10\approx 1.35\), and the mean function is \(\mu (t) = 0\). All functions are built on the closed interval [0, 1]. On each curve, 10 observations are generated at time points randomly selected from 101 equally spaced points on [0,1]. The spatial correlation between fPC scores is generated by anisotropy Matérn model in equation (2.11). In addition, as indicated in Subsect. 3.4, we estimate Matérn parameters over a list of nested \(\left\{ \varvec{\Delta }_i\right\} _{i=1}^m\)’s. In the one-dimensional case, we examine the following list: \(\{\{\varvec{\Delta }_i\}_{i=1}^m\}_{m=1}^{20} = \{(0,1),\ldots ,(0,m)\}_{m=1}^{20}\). In the two-dimensional case, the entire set of \(\varvec{\Delta }\) used for estimation is defined as \(\{\varvec{\Delta }_i\}_{i=1}^{24} =\) {(1,0), (1,1), (0,1), (1,−1), (2,0), (2,1), (2,2), (1,2), (0,2), (1,−2), (2,−2), (2,−1), (3,0), (3,1), (3,2), (3,3), (2,3), (1,3), (0,3), (1,−3), (2,−3), (3,−3), (3,−2), (3,−1)}. Estimation is performed over \(\{\varvec{\Delta }_1,\ldots ,\varvec{\Delta }_{m+4}\}_{m=1}^{20}\). Final estimates are derived as the 20 % (each side) trimmed mean across m and the order of \(\varvec{\Delta }_i\) does not have meaningful impact on the estimates.
6.1 Model estimation
We examine the estimation of SPACE model in both one-dimensional and two-dimensional cases. In one-dimensional case, we are interested in the estimation of \(\zeta \), whereas the estimation of \(\alpha \) is our main focus in two-dimensional case. Results are summarized in Tables 1 and 2. Note the first-order derivative of Matérn correlation with respect to \(\zeta \) is flattened as the correlation approaches 1. Thus, more extreme large estimates of \(\zeta \) are expected, which lead to the positive skewness observed in Table 1. We also look at the estimation performance in the correlation space. Specifically, we examine the estimated correlation at \(\varvec{\Delta }^{\mathrm{eval}} = (0,1)\). Estimation is more difficult in two-dimensional case where two more parameters \(\alpha \) and \(\delta \) need to be estimated. In general, estimates based on more significant fPCs have better quality in terms of root-mean-squared error (RMSE). To assess the curve reconstruction performance, consider a grid of time points for function evaluation \({\mathbf{t}^{\mathrm{eval}}} = (\mathbf{t}^{\mathrm{eval}}_1,\mathbf{t}^{\mathrm{eval}}_2,\ldots ,\mathbf{t}^{\mathrm{eval}}_n)\). Let \(\mathrm{{Err}} = \dfrac{1}{nN}\sum _{i=1}^N\sum _{j=1}^n(X_i(\mathbf{t}^{\mathrm{eval}}_j) - {\hat{X}}_i(\mathbf{t}^{\mathrm{eval}}_j))^2\) be the curve reconstruction error. We have reported the RMSE of the reconstructed curves under the different scenarios, both under SPACE and PACE and noted that the overall improvement in RMSE for SPACE over PACE is between 5 and 10 %. Additionally, it has also defined the reconstruction improvement (IP) as \(\mathrm{IP} = \log (\mathrm{{Err}}_{{\textsc {PACE}}}\big /{\mathrm{Err}}_{{\textsc {SPACE}}})\). Out of 100 simulated datasets, we calculate the percentage of IP greater than 0. The improvement of SPACE over PACE is more prominent in scenarios with larger noise and higher spatial correlation. It is easy to show that when \(\sigma = 0\) and the number of eigenfunctions is greater than the maximum number of observations per curve, SPACE produces exactly the same reconstructed curves as PACE. If noise is large, information provided by each curve itself is more contaminated by noise relative to neighboring locations which provide more useful guidance in curve reconstruction. As spatial correlation increases, observations at neighboring locations are more informative.
6.2 Hypothesis test
We evaluate the hypothesis tests proposed in Sect. 5. Separability and isotropy tests are implemented based on simulated datasets on one-dimensional and two-dimensional locations, respectively. In each test, 100 curves are created in each dataset and 25 datasets are generated. We first focus on separability test by examining different statistics: absolute difference in \(\zeta _1\) and \(\zeta _2\), difference of spatial correlation at \(\varvec{\Delta }^{\mathrm{eval}} = (0,1)\), and absolute difference of spatial correlation at \(\varvec{\Delta }^{\mathrm{eval}} = (0,1)\). Next, we test the isotropy effect assuming separability and the test statistics is simply \(\hat{\alpha }\). Each alternative test is evaluated at a single set of parameters. All settings are described in Tables 3 and 4. With nominal size set to 5 %, the two-sided empirical sizes and powers are summarized in Tables 3 and 4. Both tests deliver reasonable sizes and powers.
7 Harvard forest data
SPACE model is motivated by the spatial correlation observed in the Harvard Forest vegetation index data described in Sect. 2.2 and observed in Liu et al. (2012). In this section, we evaluate the SPACE model and isotropy test on the Enhanced Vegetation Index (EVI) at Harvard Forest, while the same dataset previously examined in Liu et al. (2012). EVI is constructed from surface spectral reflectance measurements obtained from moderate resolution imaging spectroradiometer onboard NASA’s Terra and Aqua satellites. In particular, the EVI data used in this work is extracted for a 25 pixel window which covers approximately an area of 134 \(\mathrm {Km}^2\). The area is centered over the Harvard Forest Long Term Experimental Research site in Petershan, MA. The data are provided at 8-day intervals over the period from 2001 to 2006. More details about the Harvard Forest EVI data can be found in Liu et al. (2012).
We first focus on verifying the directional effect observed in the second fPC scores through the proposed isotropy test. The Harvard Forest EVI data are observed over a dense grid of regularly spaced time points. EVI observed at each individual location is smoothed using regularization approach based on the expansion of saturated Fourier basis. Specifically, the sum of squared error loss and the penalization of total curvature is minimized. Let \(x_i(t)\) be the smoothed EVI at the ith location and \(\theta _k(t)\) be the kth basis function. Define the \(N\times 1\) vector-valued function \(\mathbf{x}(t) = (x_1(t),\ldots ,x_N(t))^T\) and the \(K\times 1\) vector-valued function \(\varvec{\theta }(t) = (\theta _1(t),\ldots ,\theta _K(t))^T\). All N curves can be expressed as \(\mathbf{x}(t) = \mathbf{C}{\varvec{\theta }}(t)\) where \(\mathbf{C}\) is the coefficient matrix of size \(N\times K\). The functional PCA problem reduces to the multivariate PCA of the coefficient array \(\mathbf{C}\). Assuming \(K < N\) and let \(\mathbf{U}\) be the \(K\times K\) matrix of eigenvectors of \(\mathbf{C}^T\mathbf{C}\). Let \({\varvec{\phi }}(t) = (\phi _1(t), \ldots , \phi _K(t))^T\) be the vector-valued eigenfunction and \(\varvec{\xi }\) be the \(N\times K\) matrix of fPC scores. Then we have \({\varvec{\phi }}(t) = \mathbf{U}^T{\varvec{\theta }}(t)\), \(\varvec{\xi }= \mathbf{CU}\) and \(\mathbf{x}(t) = {\varvec{\xi }}{\varvec{\phi }}(t)\).
The smoothing method is described in Liu et al. (2012) and more general introduction can be found in Ramsay and Silverman (2005). Since we have dense observations of the Harvard Forest Data, instead of smoothing the covariance and cross-covariance surfaces, we pre-smooth the data and obtain fPC scores directly. The anisotropy Matérn parameters are then estimated from the covariance of these scores. The testing procedure described in Sect. 5.2 is then applied to this modified method of finding Matérn parameters. Following the proposed procedure, we only resample the second fPC scores as they present potential anisotropy effect. For simplicity, we construct noiseless hypothesized curves so re-smoothing is not needed. The null distribution of the estimated anisotropy angle \(\alpha \) is shown in Fig. 3. The result suggests the rejection of isotropy effect and confirms the diagonal pattern in fPC scores.
500 sets of hypothesized curves are constructed in the isotropy test. In the plot on the left, upper and lower 2.5 % percentiles of the null distribution are indicated by green vertical lines. The observed anisotropy angle is 43 degree indicated by the red vertical line. The plot on the right shows the distribution of \(\mathrm{IP} = \log (\mathrm{{Err}}_{{\textsc {PACE}}}\big /\mathrm{{Err}}_{{\textsc {SPACE}}})\). To assess the gap-filling performance of SPACE versus PACE, 100 sparse samples are created for each year. In each sample, 5 observations are randomly selected per curve. In the calculation of reconstruction error, the smoothed EVI based on dense observations that is used in the isotropy test serves as the underlying true process. (Color figure online)
Next, we apply SPACE framework to the gap-filling of Harvard Forest EVI data. To that end, we create sparse samples by randomly selecting 5 observations from each location and each year. 100 sparse samples are created. To make the estimation and reconstruction of EVI across 625 pixels more computationally tractable, both SPACE and PACE are performed in each year, respectively. The distribution of IP in year 2005 is summarized in Fig. 3. 100 % of the 100 samples show improved reconstruction performance using SPACE. Similar results are also achieved for other years. By incorporating spatial correlation estimated from EVI data, better gap-filling can be achieved.
8 Conclusion
Much of the literature in functional data analysis assumes no spatial correlation or ignoring spatial correlation if it is mild. We propose the spatial principal analysis based on conditional expectation (SPACE) to estimate spatial correlation of functional data, using nonparametric smoothers on curves and surfaces. We find that the leave-one-bin-out cross-validation based on binned data performs well in selecting bandwidth for local linear smoothers. Empirical spatial correlation is calculated as the ratio of eigenvalues of cross-covariance and covariance surfaces.
A parametric model, Matérn correlation augmented with anisotropy parameters, is then fitted to empirical spatial correlations at a sequence of spatial separation vectors. With finite sample, estimates are better for separable covariance than nonseparable covariance. The fitted anisotropy Matérn parameters can be used to compute the spatial correlation at any given spatial separation vector and thus are used to reconstruct trajectories of sparsely observed curves. We present a series of asymptotic results which demonstrate the consistency of model estimates. In particular, the same convergence rate for correlated case as that of i.i.d. case is derived assuming mild spatial correlation structure.
This work compares with the work in Yao et al. (2005) where curves are assumed to be independent. We show that by incorporating the spatial correlation, reconstruction performance is improved. It is observed that the higher the noise and true spatial correlations, the greater the improvements. We demonstrate the flexibility of SPACE model in modeling the separability and anisotropy effect of covariance structure. Moreover, two tests are proposed as well to explicitly answer if covariance is separable and/or isotropy. Reasonable empirical sizes and powers are obtained in each test.
In our application of the SPACE method to Harvard Forest EVI data, we confirm the diagonal pattern observed in the second fPC scores through a slightly modified version of the proposed isotropy test. Moreover, we demonstrate that by taking into account explicitly the spatial correlation, SPACE is able to produce more accurate gap-filled EVI trajectory on average.
9 Supplementary materials
The supplementary materials provide a detailed proof of Theorem 1* generalizing the consistency results from Yao et al. (2005). It expands on the conditions needed for the genralized results and shows the asymptotic properties of SPACE model components. The R-codes for implementing all the analyses and simulations described in the paper are available at http://www.stats.gla.ac.uk/~sray/software/.
References
Abramowitz, M., Stegun, I.: Handbook of Mathematical Functions. Dover, New York (1970)
Abramowitz, M., Stegun, I.A., et al.: Handbook of mathematical functions. Appl. Math. Ser. 55, 62 (1966)
Aston, J.A., Pigoli, D., Tavakoli, S.: Tests for separability in nonparametric covariance operators of random surfaces. arXiv preprint arXiv:1505.02023 (2015)
Avriel, M.: Nonlinear Programming: analysis and Methods. Courier Corporation, New York (2003)
Baladandayuthapani, V., Mallick, B.K.: Young Hong, M., Lupton, J.R., Turner, N.D., Carroll, R.J.: Bayesian hierarchical spatially correlated functional data analysis with application to colon carcinogenesis. Biometrics 64(1), 64–73 (2008)
Banerjee, S., Johnson, G.A.: Coregionalized single-and multiresolution spatially varying growth curve modeling with application to weed growth. Biometrics 62(3), 864–876 (2006)
Bowman, A. W. and Azzalini, A.: R package sm: nonparametric smoothing methods (version 2.2-5), University of Glasgow, UK and Universit\({\grave{a}}\) di Padova, Italia. http://www.stats.gla.ac.uk/adrian/sm, http://azzalini.stat.unipd.it/Book_sm (2013)
Cressie, N.: Statistics for Spatial Data. Wiley, New York (2015)
Diggle, P.J., Ribeiro Jr., P.J., Christensen, O.F.: An Introduction to Model-Based Geostatistics, Spatial Statistics and Computational Methods, pp. 43–86. Springer, New York (2003)
Gromenko, O., Kokoszka, P., Zhu, L., Sojka, J.: Estimation and testing for spatially indexed curves with application to ionospheric and magnetic field trends. Ann. Appl. Stat. 6(2), 669–696 (2012). doi:10.1214/11-AOAS524
Hall, P., Müller, H., Wang, J.: Properties of principal component methods for functional and longitudinal data analysis. Ann. Stat. 34(3), 1493–1517 (2006)
Hurvich, C.M., Simonoff, J.S., Tsai, C.-L.: Smoothing parameter selection in nonparametric regression using an improved akaike information criterion. J. R. Stat. Soc. 60(2), 271–293 (1998)
James, G., Sugar, C.: Clustering for sparsely sampled functional data. J. Am. Stat. Assoc. 98(462), 397–408 (2003)
Li, P.-L., Chiou, J.-M.: Identifying cluster number for subspace projected functional data clustering. Comput. Stat. Data Anal. 55(6), 2090–2103 (2011)
Li, Y., Hsing, T.: Uniform convergence rates for nonparametric regression and principal component analysis in functional and longitudinal data. Ann. Stat. 38(6), 3321–3351 (2010)
Li, Y., Wang, N., Hong, M., Turner, N.D., Lupton, J.R., Carroll, R.J.: Nonparametric estimation of correlation functions in longitudinal and spatial data, with application to colon carcinogenesis experiments. Ann. Stat. 35(4), 1608–1643 (2007)
Liu, C., Ray, S., Hooker, G., Friedl, M.: Functional factor analysis for periodic remote sensing data. Ann. Appl. Stat. 6(2), 601–624 (2012)
Mateu, J.: Spatially correlated functional data, in ‘Spatial2 conference: spatial data methods for environmental and ecological processes, Foggia (IT), 1–2 September 2011’, IT (2011)
Menafoglio, A., Petris, G.: Kriging for hilbert-space valued random fields: the operatorial point of view. J. Multivar. Anal. 146, 84–94 (2016). doi:10.1016/j.jmva.2015.06.012
Paul, D., Peng, J.: Principal components analysis for sparsely observed correlated functional data using a kernel smoothing approach. Electron. J. Stat. 5, 1960–2003 (2011)
Peng, J., Paul, D.: A geometric approach to maximum likelihood estimation of the functional principal components from sparse longitudinal data. J. Comput. Graph. Stat. 18(4), 995–1015 (2009)
R Development Core Team.: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna (2010)
Ramsay, J.O., Silverman, B.W.: Functional Data Analysis, 2nd edn. Springer, New York (2005)
Rice, J., Silverman, B.: Estimating the mean and covariance structure nonparametrically when the data are curves. J. R. Stat. Soc. Ser. B (Methodol.) 53(1), 233–243 (1991)
Ruppert, D., Carroll, R.J., Gill, R., Ripley, B.D., Ross, S., Stein, M., Wand, M.P., Williams, D.: Semiparametric Regression. Cambridge University Press, Cambridge. https://cds.cern.ch/record/997686 (2003)
Staicu, A., Crainiceanu, C.M., Reich, D.S., Ruppert, D.: Modeling functional data with spatially heterogeneous shape characteristics. Biometrics 68(2), 331–343 (2012)
Staicu, A.M., Crainiceanu, C.M. and Carroll, R.J. (2010) Fast methods for spatially correlated multilevel functional data. Biostatistics 11(2), 177–194. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2830578/pdf/kxp058.pdf
Yao, F., Lee, T.: Penalized spline models for functional principal component analysis. J. R. Stat. Soc. Ser. B 68(1), 3 (2006)
Yao, F., Müller, H., Clifford, A., Dueker, S., Follett, J., Lin, Y., Buchholz, B., Vogel, J.: Shrinkage estimation for functional principal component scores with application to the population kinetics of plasma folate. Biometrics 59(3), 676–685 (2003)
Yao, F., Müller, H., Wang, J.: Functional data analysis for sparse longitudinal data. J. Am. Stat. Assoc. 100(470), 577–590 (2005)
Zhou, L., Huang, J.Z., Martinez, J.G., Maity, A., Baladandayuthapani, V., Carroll, R.J.: Reduced rank mixed effects models for spatially correlated hierarchical functional data. J. Am. Stat. Assoc 105(489), 390–400. http://EconPapers.repec.org/RePEc:bes:jnlasa:v:105:i:489:y:2010:p:390-400 (2010)
Acknowledgments
The authors gratefully acknowledges the support of the National Science Foundation under Grant No 0934739.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Liu, C., Ray, S. & Hooker, G. Functional principal component analysis of spatially correlated data. Stat Comput 27, 1639–1654 (2017). https://doi.org/10.1007/s11222-016-9708-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11222-016-9708-4