Abstract
Automated extraction of coronary arteries is an essential process in the diagnosis of treatment for coronary artery disease (CAD) with computer assistance. Accurately outlining the coronary artery is difficult when using X-ray coronary angiography (XCA) because of the low signal-to-noise ratio and the presence of interfering background structures. In this paper, a new approach for segmenting vessels in angiograms is presented, specifically designed to tackle the difficulties arising from non-uniform illumination, artifacts, and noise present in angiographic images. The proposed method employs an edge-based tracking tool to generate an initial probability map for segmentation. A segmentation method based on coronary vessel tracking is presented for finding the border and centerline of the vessel. The proposed method is designed based on two main components: preprocessing and tracking. In the preprocessing stage, a guided filter and edge-sharpening algorithms are used to enhance the features of the original image. In the tracking stage, an initial point is selected, and using the Gaussian property, a semi-circle operator is applied to track the line perpendicular to the vessel. The proposed method demonstrated remarkable performance in terms of sensitivity and specificity, achieving values of 86.93 and 99.61, respectively. Additionally, the method achieved an accuracy rate of 97.81. Notably, the proposed method outperformed existing state-of-the-art segmentation methods, as indicated by its higher dice score. These impressive results signify a significant advancement in the field of vessel segmentation, highlighting the effectiveness and superiority of the proposed approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Coronary artery disease (CAD) is the most prevalent type of heart disease and remains the leading cause of global mortality, affecting approximately 330 million individuals and resulting in the loss of 17.9 million lives annually [1, 2]. X-ray coronary angiography (XCA) is considered the gold standard for diagnosing CAD [3]. It involves the administration of contrast dye into the coronary vessels, followed by the visualization of its flow through 2D projections of the vessel structure. XCA aids clinicians in identifying potential stenoses, assessing their severity visually, and determining the most appropriate interventional treatments [4]. However, visual stenosis assessment based on XCA images is prone to unreliability. It tends to overestimate severe blockages while underestimating milder ones [5, 6], and it exhibits high variability among different observers and even within the same observer [7, 8]. To provide a more objective evaluation of the lumen diameter, quantitative coronary angiography (QCA) was introduced [9]. QCA enables semi-automatic analysis of XCA images [10, 11]. Vessel segmentation is a crucial step in QCA as it is necessary for calculating the percentage of arterial stenosis.
Extracting blood vessels from XCA images is a critical initial step in developing a computer-assisted diagnostic and therapeutic framework, which encompasses tasks such as stent placement [12]. Due to the time-consuming and costly nature of manually annotating vessel structures, there has been significant research on automatic or semi-automatic computer-assisted segmentation methods [13,14,15]. However, these methods face challenges when it comes to vessel segmentation, including the presence of low contrast, noise, and artifacts in angiography images, and the complexity of vessel structures, such as multi-scale structures, bifurcation structures, and overlapping structures. As a result, achieving precise vessel segmentation from XCA images continues to be a demanding task, requiring ongoing innovations and advancements in image processing and computational techniques.
The organization of the rest of this paper is as follows: In Sect. 2, the related works are explained. The proposed method is presented in Sect. 3. Section 4 explains the experimental results, and Sect. 5 concludes the paper.
2 Relatd Studies
In recent years, several researchers have made significant progress in developing effective methods for vessel segmentation. The most used Hessian matrix-based method, proposed by Frangi et al [16], involves designing a multi-scale second-order derivative filter to characterize vessel structures. This approach computes the eigenvalues and eigenvectors of the Hessian matrix. However, to address the non-uniform response observed in Frangi’s method, Jerman et al. [17] introduced an enhanced technique relying on the Hessian matrix for improving angiographic images. This method introduces a novel vesselness function designed to accurately represent the elliptical cross-sections inherent in vessel structures.
Several segmentation frameworks have been proposed that incorporate vessel features for vessel segmentation [18,19,20,21]. Unsupervised methods for automatic coronary vessel segmentation can be broadly categorized into tracking-based, model-based, and filter-based approaches [22].
Tracking-based methods [23] involve selecting seed points on vessel edges or centerlines and iteratively searching for nearby vessel edges or centerlines. These methods estimate the vessel direction and take steps in that direction to find new edges. While tracking-based methods maintain continuity in the segmentation of the vessel tree structure, they heavily rely on the quality of the initial seed points and may struggle to handle background elements adjacent to vessels. Certain tracking methods [24,25,26] have constructed the segmentation pipeline by incorporating prior information derived from structures like centerlines or surfaces.
Model-based methods [27,28,29,30,31,32], utilize deformable models or region growing techniques to evolve the segmentation towards vessel-background boundaries based on energy functions. These methods apply forces and constraints defined by the energy functions and may use similarity functions with threshold parameters as growing conditions. Model-based methods also require initial seeds for segmentation, making them sensitive to initialization. While they often achieve good segmentation continuity for the vessel tree structure, they may struggle with confounding elements in the background.
Filter-based methods [33] employ various filters for tasks such as non-uniform background intensity balancing, suppression of irrelevant structures, noise reduction, and vessel enhancement. The filtered images are then processed using thresholding techniques to generate segmentation masks. Filter-based methods are easy to implement and can mitigate illumination issues. They are commonly used as preprocessing steps in both supervised and unsupervised methods for automated coronary vessel segmentation [32, 34,35,36,37,38,39]. However, they are typically insufficient on their own as they can be sensitive to background structures and may not perform well on vessel junctions and bifurcations [40]. In accordance with the findings from studies on the Hessian matrix, several scholars have introduced a multidirectional filter bank. This implementation results in a heightened filter response along the vessel direction within the feature map [38, 41, 42]. The primary objective of the multi-directional filter bank is to diminish background noise and augment the response of vessels. As X-ray angiography images constitute a temporal series, certain investigations have aimed to maximize temporal information. This has been achieved through matrix decomposition, employed to eliminate background elements and yield a more precise blood vessel tree [30, 43, 44]. Nevertheless, it is noteworthy that matrix decomposition, being an iterative process, entails a substantial amount of time for high-dimensional matrix decomposition.
Deep learning methods have made considerable strides because of its ability to independently learn powerful feature representations from dataset and have been used for segmentation in numerous applications [45]. Moreover, The essential reliance of deep learning algorithms on data underscores the crucial requirement for robust datasets to demonstrate optimal performance [46]. Ronneberger et al. [47] proposed a U-shaped network architecture known as UNet. This architecture consists of an encoding path, a decoding path, and skip-connection paths that facilitate the transmission of feature maps from the encoding path to the decoding path. UNet has demonstrated strong performance across various medical applications. Numerous beneficial structures have been suggested to enhance the efficacy of deep learning models, including normalization [48,49,50,51], residual blocks [52, 53], dense blocks [54, 55], inception blocks [56, 57], dilated convolution [58], feature aggregation modules [59], spatiotemporal super-resolution frameworks [60,61,62], encoder-decoder architectures [63, 64], and more. Present research on 2D vessel segmentation methods is predominantly concentrated on retinal vessel segmentation. Zhu et al. [65] employed the Pyramid Scene Parsing Network, which was originally proposed by [66], for the segmentation of coronary vessels. They utilized the network’s structure to incorporate features from multiple scales through pyramid pooling and applied transfer learning to prevent overfitting on a limited training dataset. Navarro et al. [67] introduced two auxiliary tasks, namely a distance map and a contour map, at the network’s end to achieve more precise segmentation. Concurrently, an increasing number of deep learning models [68,69,70,71,72] are being employed for tasks involving the segmentation of volume data.
When it comes to supervised methods for coronary artery segmentation, the focus may be primarily on the major coronary arteries, as they are of greater concern to clinicians, rather than the entire arterial tree. While network-based supervised methods have certain limitations, such as overfitting with small training sets, reduced interpretability compared to unsupervised filter-based methods, and challenges in ensuring connectivity within predicted masks, they still offer advantages such as requiring less manual input and demonstrating greater robustness in distinguishing background structures like catheters and spines, in comparison to unsupervised methods.
This paper introduces a segmentation method that utilizes coronary vessel tracking to identify the border and centerline of the vessel. The method consists of two primary components: preprocessing and tracking. During the preprocessing stage, the original image is subjected to feature enhancement through the application of a guided filter and edge-sharpening algorithms. In the tracking stage, an initial point is chosen, and a semi-circle operator is employed, leveraging the Gaussian property, to track the line that is perpendicular to the vessel.
3 Proposed Method
This technique segments coronary vessels by tracking their border and centerline using two main components: preprocessing and tracking. Preprocessing enhances the original image using guided filtering and edge-sharpening algorithms. In the tracking phase, an initial point is chosen, and a semi-circle operator based on the Gaussian property is used to track the vessel’s perpendicular line The diagram of the proposed method is shown in Fig. 1.
3.1 Pre-Processing
The preprocessing stage focuses on reducing noise and enhancing vessel edges without extensive modifications to the image. Additional filters are avoided to preserve the Gaussian property of the vessel in the cross-sectional line. The brightness level of the image is adjusted between 0 and 255, and image noise is reduced using a guided filter described in Sect. 3.1.1. The Unsharp-Masking method described in Sect. 3.1.2 is employed to enhance edge visibility.
3.1.1 Guided Filter
In [73], a filter is designed to improve the quality of images. The guided filter serves as an edge-preserving filter that transfers the characteristics of the original image to the output image based on a guide image. The guide image can be the same as the input image or a different image. Considering I as the guide image and p as the input image for quality enhancement, the output image (q) is obtained using Eq. 1.
Where i and j are pixel indexes, and the filter kernel \(W_{ij}\) is a function of the guide image I, which is independent of the input image. The guided filter is a linear filter based on the input image. It can be assumed that q is a linear transformation of I within a window \(\omega _k\) centered at the pixel k, and \(a_k\) and \(b_k\) are constant linear coefficients within the window.
Accordingly, the images can be divided into overlapping windows with a radius of r. It can be established that within each window, q exhibits edge-preserving behavior only if it has edges. In other words, a model can be proposed to suppress the additional and unwanted components n of the input image from it like noise/textures:.
To preserve the structure of the input image and maintain the edges, the difference between p and q should be minimized. Therefore, in Eq. 4, the cost function within the window \(\omega _k\) should be minimized. The cost function represents the discrepancy between the input and guided images.
Here \(\epsilon\) is a regularization parameter penalizing large \(a_k\). The energy function is minimized concerning the values of \(a_k\) and \(b_k\) in Eqs. 5 and 6. By minimizing the energy function, the optimal values of \(a_k\) and \(b_k\) are obtained, which help minimize the discrepancy between p and q within the window.
In these equations, \(\mu _k\) and \(\sigma _k^2\) represent the mean and variance of I within the window \(\omega _k\), respectively. \(|\omega |\) denotes the number of pixels in the window \(\omega _k\). \(p_k=1/|\omega | \sum _{i\ \in \omega _k} p_i\) is the mean of p within the window \(\omega _k\). With the known values of coefficients (\(a_k\) and \(b_k\)), the output image can be obtained based on Eq. 2 (Fig. 2).
The process of obtaining the filtered image using the guided image [73]
3.1.2 Unsharp-Masking Edge Enhancement Filter
The Unsharp-Masking algorithm is a technique for sharpening digital images [74, 75]. In this algorithm, a smoothed version (\(f_{smooth}\)) of the original image (f) is subtracted to obtain the edge image (g). In the next step, the sharpened image (\(f_{sharp}\)) is obtained by adding the original image multiplied by a coefficient of the edge image, which essentially emphasizes the high-frequency components of the original image. Figure 3 illustrates the steps of creating a sharp signal using this method and the effects when applied to angiography images. As shown in the figure, the details and edges are better revealed in the image where this algorithm is applied. In Eq. 7, \(G_\sigma\) refers to a Gaussian filter with standard deviation \(\sigma\). In the below equations, the standard deviation is set to 2, and the gain (K) is set to 1.
3.2 Edge-Based Tracking Method
This section presents a dynamic algorithm for vessel tracking, which begins from an initial point (O) selected by the user. To handle branch points and vessel crossings, the algorithm creates a list to store these points. Additionally, another list is created to store detected vessel points along with their conditions, such as vessel radius and intensity level. The algorithm utilizes a semicircular operator to track and identify edge points of the vessel. Vessel characteristics are extracted from the vicinity of this operator to determine the type of location, including branch points, vessel crossings, end points, and normal vessels. This process is repeated until the entire vessel is traversed.
3.2.1 Overall Process of the Tracking Algorithm
Assuming in Figs. 4 and 5, the points corresponding to the i-th iteration are marked. Initially, a circle with a radius of R is considered around the center point \(P_c^i\). This circle is divided into two parts by a diameter passing through points \(P_L^i\) and \(P_R^i\).
\(\bullet\) Calculation of the angle between two previous points: To calculate the angle of the line between points \(P_L^i\) and \(P_R^i\), the slope of the line passing through these two points is first calculated. Then, the angle \(\theta _i\) corresponding to the slope m is determined.
The slope of the line is obtained using Eq. 11. In this equation, (\(Y_L^i\),\(X_L^i\)) represents the coordinates of the left point, and (\(Y_R^i\),\(X_R^i\)) represents the coordinates of the right point on the vessel.
\(\bullet\) Determining the direction of the semicircle: To select one of the two sides of the semicircle, the previous detected point positions are used (Fig. 5). For this purpose, the direction of the operator’s movement to detect the previous points is determined, and a corresponding semicircle is chosen based on that direction. Four directions are considered for the movement of the vessel, as shown in Figure 5. Based on the positions of the previous points, the new direction of the semicircle is determined in the i-th iteration according to 1 (Table 1).
\(\bullet\) Detection of the boundary of the vessel: In Fig. 6, the pixels along the linear path through the vessel exhibit a shape resembling the inverse Gaussian function. However, this shape is not easily analyzable in all cases, making it challenging to determine the vessel boundary from such a plot. The proposed method in this work introduces three approaches to predict the points on the vessel edge.
The selection of a suitable boundary location is necessary to remove the irregularities present in the vessel characteristic. For this purpose, each component in the characteristic is averaged with its neighbors in two stages. Then, the locations of the valleys in the vessel characteristic are identified, and the deepest valley Vmin is considered as the center point of the vessel at coordinates (\(x_v\),\(y_v\)).
In the first method, the characteristic array of the vessel is divided into two parts from the valley point, and the Otsu thresholding method is applied separately to each part to determine the threshold values \(T_{left}\) and \(T_{right}\). From the \(V_min\) point, the vessel’s characteristic is traversed towards the left and right, and the first value that exceeds the threshold is chosen as the vessel boundary, determining the two points \(p_1\) and \(p_2\).
If the first method fails to find points around the valley, a window with dimensions of 30*30 centered at (\(x_v\), \(y_v\)) is considered. The brightness of this window is adjusted, and an edge detection filter is applied to it. Then, the first edges found on both sides of the valley are selected as the initial boundary points \(p_1\) and \(p_2\). Sometimes it may not be possible to obtain an edge. In this case, the width of the valley is halved, and we move half of the valley width on both sides of the valley to determine the values of \(p_1\) and \(p_2\). If all three methods fail to find points on the vessel boundary, the boundary failure flag (\(P_{err}\)) is activated and indicates that the vessel has reached its end.
When the two boundary points are obtained, the midpoint of these two points, Pcenter, is recognized as the centerline point of the vessel.
The left and right boundary points (\(p_1\) and \(p_2\)) divide the space surrounding the vessel into three parts: background 1, background 2, and the vessel itself. In this work, a distinction is made between the backgrounds on both sides of the vessel. This differentiation is due to the potential difference in brightness levels between the two sides. The average value of each part is calculated and stored separately.
\(\bullet\) Point validity assessment: In this stage, the validity of the selected point is evaluated from multiple perspectives. If the selected point is not acceptable in any of the evaluations, the \(F_{BE}\) (End of Branch) flag is activated to exit the algorithm. The reasons that activate the \(F_{BE}\) flag are listed below:
- \(P_{err}\) flag activation.
- Insufficient or excessive distance between two points, \(p_1\) and \(p_2\): The geometric distance between the two points is calculated, and if it is less than 3 pixels or greater than 30 pixels, the \(F_{BE}\) flag is activated.
- Going out of the image: If the coordinates of either \(p_1\) or \(p_2\) are outside the image boundaries.
- Insufficient contrast with the background: Even in non-vessel areas (Fig. 6), there may be valleys. To identify these cases, we examine the average difference between the vessel’s brightness level and the background average. If the average brightness level of the vessel differs by less than 6 from the background average, that region is not considered a vessel.
- Failure to pass the Hessian filter: In some cases where the obtained points are not on the vessel, by considering a small window around the \(P_{center}\) and comparing the sum of vessel metrics for all pixels in the window with the product of the image’s Hessian filter threshold and the number of pixels in the window, if the sum is lower, the point is not a vessel.
- Encounter with a repeated point: With each iteration of vessel tracking and reaching two points, \(p_1^i\) and \(p_2^i\), in the i-th iteration, and two \(p_1^{i-1}\) and \(p_2^{i-1}\) points in the 1-i iteration, if both iterations belong to the same vessel branch, a quadrilateral can be drawn to represent the vessel within the distance of these two point sets. This quadrilateral can be used to obtain a cover at each stage and update it. If, during edge tracking, the algorithm encounters an area that has been previously visited, it stops in the current branch. If the \(F_{BE}\) flag is activated due to any of the six mentioned reasons, the algorithm stops in the current branch and continues in another branch. This process continues until all branches are encountered.
\(\bullet\) Detection of branch points or crossing of two vessels: In vessel tracking, branching and vessel intersections can occur. Our method detects branch points during each iteration. If a branch point is found, its central point is stored, and the algorithm continues from the next point in each branch. Detection relies on analyzing vessel characteristics. Valleys in the vessel’s linear characteristic indicate branch points. By examining characteristics at these points, additional valleys become evident, representing vessels in the image. Our proposed method detects branch points using these observations.
All valleys with a level difference greater than 8 in the characteristic, except for the deepest valley examined in the boundary determination section, are placed in a list, and the presence of an edge point on both sides is examined. Initially, for each valley, a point for the left edge and a point for the right edge are calculated by moving from the minimum point of the valley \((V_{min}^m\)) towards the left and right by half the width of the valley. These points are denoted as \(p_{Lw}^m\) and \(p_{Rw}^m\), respectively. Similarly, as described in the boundary identification section, two threshold levels, \(T_{left}^m\) and \(T_{right}^m\), are determined for each valley, and on the left and right sides of each valley, the first point that exceeds these threshold values is recognized as \(p_{Lt}^m\) or \(p_{Rt}^m\).
In this method, the vessel characteristic is smoothed initially. Valleys exceeding a difference of 8 compared to the higher level are selected. Two boundary calculation methods are applied to determine the vessel’s sides. The results are combined for further analysis.
In this stage, the results of the two methods for each boundary are combined to select the boundary between the obtained boundaries. For determining the left boundary (\(p_L^m\)), the boundary between the boundaries obtained by the two methods (\(p_{Lw}^m\) and \(p_{Lt}^m\) ) is selected as the left boundary if it is farther away than \(V_{min}^m\), in other words, more towards the left. Similarly, for determining the right boundary (\(p_R^m\)), the boundary between the two methods that is more towards the right is selected as the right boundary. Then, the midpoint between these two boundaries (\(p_{center}^m\)) is calculated and used as the starting point of a new branch, which is stored. To store the starting points of new branches, the central point of the vessel (\(p_{center}^m\)), the edges (\(p_{Lw}^m\) and \(p_{Lt}^m\) ) the characteristics of the two points before branch detection (\(p_{center}^i\) and \(p_{center}^{i-1}\)), the angle of the line perpendicular to the vessel at the central point \(p_{center}^m\) (\(\theta _L\)), and the number of the detected point (i) are added to the branch point list.
To calculate the new radius for considering the circle, the value of the radius changes based on the size of the current location’s radius. The new circle radius is calculated as follows:
In equation 12, the value of R represents the current radius of the circle, while \(R_{vessel\ }\)corresponds to half the distance between the two detected boundary points of the vessel. The offset is a parameter used to adjust the circle’s radius, and the optimal outcome is obtained when the offset is set to 1.
In this section, all the information obtained during the detection of each point on the vessel is stored in a list called "vessel points." This list includes the coordinates of the left and right points of the vessel (\(x_L^m\),\(x_R^m\), \(y_L\), \(y_R\)), their center, the average background intensity, and the status of the vessel, which indicates the starting and ending points of the vessel.
3.2.2 Starting From the Initial Point
In the initial step, a circle with a radius of R is centered around point O. A perpendicular line to the vessel passing through point O is determined to divide the circle into two semicircles. In the method that is employed to find this perpendicular line, the maximum vessel radius, r, is assumed. The circle with a diameter of 2r is rotated around point O, with each rotation being one degree. The intensity values of the vessel are stored in an array, and their average is computed (Fig. 7). As the vessel typically exhibits lower brightness compared to the background region, the line with the highest average brightness level is considered the perpendicular line to the vessel. Initially, the first semicircle is selected, and the tracking process commences from this point.
Next, the brightness characteristics of the points along the semicircle are analyzed. Figure 4 demonstrates the brightness graph of the semicircle operator for four consecutive operators. This graph reveals that the brightness levels form a shape resembling an inverse Gaussian curve, which aids in identifying the vessel boundary. Furthermore, Fig. 8 showcases the vessel traversal process and the sequence in which the algorithm traverses the detected branching sub-branches. It is apparent from this image that the sub-branches originating from earlier branching points in the initial point list are traversed first.
3.2.3 Binary Vessel Map Formation
The last phase of the suggested segmentation algorithm involves converting the probability map generated for the vessel region into a binary segmentation mask. To achieve this, a threshold is applied to the values within the final probability map. Pixels with probabilities surpassing the threshold are designated as part of the vessel region.
The proposed method utilizes an adaptive approach to determine an appropriate threshold for each image. To achieve this, the threshold is initially set to a small value of \(\epsilon\), such as \(\epsilon = 0.001\). Using this initial threshold, the largest connected component is tracked within the binary mask. Subsequently, the threshold is gradually increased, resulting in a new binary mask. As the threshold increases, fewer points are identified as part of the vessel region. The threshold increment stops when less than 80% of the points with a probability greater than 0.5 are included in the largest connected component. The obtained threshold value is then chosen for generating the final segmentation mask. This adaptive threshold selection process is employed to preserve the connectivity of the extracted vessel regions.
4 Results and Discussion
4.1 Evaluation Metrics
The performance of the proposed method is evaluated on a dataset consisting of X-ray angiography images acquired using the Siemens Artis Zee Floor Fluoroscopy system. The dataset consists of 44 grayscale images, each with dimensions of 512 \(\times\) 512 pixels. Expert-annotated ground truth is provided for segmentation evaluation.
The evaluation of the proposed vessel extraction method in X-ray angiograms involves comparing it to state-of-the-art algorithms.
The evaluation metrics used for comparison are defined by Eqs. 13,14,15 and 16:
True Positive (TP): number of pixels correctly labeled as vessel in both the ground truth and the produced segmentation mask.
False Negative (FN): number of vessel pixels missed by segmentation mask.
False Positive (FP): number of pixels that are incorrectly classified as vessel.
True Negative (TN): number of background pixels accurately labeled as background by segmentation mask.
4.2 Performance of the Proposed Method
The performance metrics are summarized in Table 2. The proposed method outperforms other algorithms in terms of segmentation accuracy, sensitivity, specificity, and Dice score. Notably, the proposed method achieves a high Dice score of 81.79% by accurately detecting vessel parts.
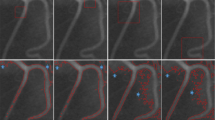
Figure 9 displays the results of applying the proposed algorithm to a dataset of X-ray angiography images. The tracking method effectively follows the main branches of the vessel and accurately identifies the main bifurcations. However, in smaller vessels, the method occasionally fails to exit the loop. Nevertheless, where the method successfully traverses, it provides a reliable estimation of the vessel boundary, as depicted in the images.
Visual results of the proposed methods, along with the available ground truth for segmentation, are presented in Fig. 10. In 10-a, the automatically detected branching points are highlighted. In 10-b, the method successfully performs orientation estimation. The displayed image in the figure contains points in all directions as illustrated in Fig. 5, demonstrating accurate determination of the orientation in the applied method.
In the realm of coronary arteries segmentation, we presented a novel approach for extraction of coronary arteries in X-ray coronary angiography (XCA) images, with a specific focus on addressing challenges related to non-uniform illumination, artifacts, and noise. The proposed method consists of two main components: preprocessing and edge-based tracking. In the preprocessing stage, guided filtering and edge-sharpening algorithms are applied to enhance the features of the original angiographic images. This step is crucial for improving the accuracy of subsequent vessel segmentation, as it helps to preserve edges and enhance contrast. The edge-based tracking method employs a dynamic algorithm starting from a user-selected initial point. This algorithm is designed to handle branch points and vessel crossings, recording them in a list for further analysis. A semicircular operator is used to locate and track the vessel’s edge points, and the characteristics extracted from the operator determine the nature of the location, such as branch points, vessel crossings, endpoints, or regular segments. It is noteworthy to emphasize that our proposed method operates as a semi-automatic approach in contrast to fully automatic machine learning methods often used in the field. While fully automatic methods, including those based on Convolutional Neural Networks (CNNs) have demonstrated effectiveness in learning complex patterns independently, our approach integrates user-guided input for enhanced control and interpretability in the segmentation process. This recognition of the semi-automatic nature of our method aligns with the ongoing discourse in medical image analysis, highlighting the importance of balancing computational precision with human expertise. The deliberate involvement of user input in selecting the initial point adds a layer of interpretability and control, allowing clinicians to influence and validate results based on their expertise. Our proposed method demonstrated remarkable performance in terms of sensitivity, specificity, and accuracy. The sensitivity and specificity values of 86.93 and 99.61, respectively, highlight the ability to accurately identify vessel regions while minimizing false positives. The overall accuracy rate of 97.81 further underscores the reliability of our approach in segmenting coronary vessels. Comparison with existing state-of-the-art segmentation methods revealed that our proposed method outperformed others, as evidenced by its higher Dice score. The Dice score of 81.79% signifies a significant improvement in accurately detecting vessel parts compared to other contemporary techniques. These findings position our method as a notable advancement in the field of coronary vessel segmentation. In practical applications, our algorithm demonstrated effectiveness in following main vessel branches and accurately identifying bifurcations. Challenges were observed in smaller vessels where the algorithm occasionally struggled to exit loops. Despite these limitations, where successful, the algorithm provided reliable estimations of vessel boundaries. Exploration of further investigations and upcoming research initiatives is currently in progress, anticipating the advancement of more efficient models for clinical applications by leveraging larger and more varied datasets in the future.
5 Conclusion
This paper introduces a robust and effective method for the segmentation of coronary vessels in X-ray angiography images, addressing challenges associated with non-uniform illumination, artifacts, and noise. The proposed technique leverages an edge-based tracking approach, incorporating preprocessing steps such as guided filtering and edge-sharpening to enhance image features. The dynamic tracking algorithm efficiently identifies vessel boundaries, branch points, and crossings, providing a comprehensive framework for vessel segmentation. The results of the evaluation on a dataset of X-ray angiograms demonstrate the superiority of the proposed method over state-of-the-art algorithms, achieving outstanding performance metrics. With a Dice score of 81.79%, sensitivity of 86.93%, and an accuracy rate of 97.81%, the proposed approach surpasses existing methods in accurately delineating coronary vessels. The comparative analysis against established techniques such as CNN and PSPnet further highlights the competitiveness and potential of the proposed method in the field of coronary vessel segmentation. In practical applications, the proposed algorithm exhibits reliability in tracking main vessel branches and accurately identifying bifurcations. While challenges may arise in smaller vessels, the overall performance of the method underscores its efficacy in providing precise vessel boundary estimations. This research contributes a significant advancement in coronary vessel segmentation, offering a compelling alternative to existing methods. The notable achievements in terms of accuracy and robustness position the proposed approach as a valuable tool in computer-assisted diagnosis of coronary artery disease. Future work may explore optimizations and adaptations for specific clinical scenarios, further validating the practical utility of this method in real-world medical applications.
Data availability
All relevant datasets used in this study will be made available upon request.
References
Wang, H., Naghavi, M., Allen, C., Barber, R., Bhutta, Z., Carter, A., et al. (2016). A systematic analysis for the global burden of disease study 2015. Lancet, 388(10053), 1459–1544.
Lipton, R., Schwedt, T., & Friedman, B. et al. (2016). Gbd 2015 disease and injury incidence and prevalence collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: A systematic analysis for the global burden of disease study 2015. Lancet 388(10053), 1545–1602.
James, S. L., Abate, D., Abate, K. H., Abay, S. M., Abbafati, C., Abbasi, N., Abbastabar, H., Abd-Allah, F., Abdela, J., Abdelalim, A., et al. (2018). Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. The Lancet, 392(10159), 1789–1858.
Tavakol, M., Ashraf, S., & Brener, S. J. (2012). Risks and complications of coronary angiography: A comprehensive review. Global Journal of Health Science, 4(1), 65.
Fleming, R. M., Kirkeeide, R. L., Smalling, R. W., Gould, K. L., & Stuart, Y. (1991). Patterns in visual interpretation of coronary arteriograms as detected by quantitative coronary arteriography. Journal of the American College of Cardiology, 18(4), 945–951.
Zhang, H., Mu, L., Hu, S., Nallamothu, B. K., Lansky, A. J., Xu, B., Bouras, G., Cohen, D. J., Spertus, J. A., Masoudi, F. A., et al. (2018). Comparison of physician visual assessment with quantitative coronary angiography in assessment of stenosis severity in China. JAMA Internal Medicine, 178(2), 239–247.
Zir, L. M., Miller, S. W., Dinsmore, R. E., Gilbert, J., & Harthorne, J. (1976). Interobserver variability in coronary angiography. Circulation, 53(4), 627–632.
DeRouen, T., Murray, J., & Owen, W. (1977). Variability in the analysis of coronary arteriograms. Circulation, 55(2), 324–328.
Serruys, P. W., Reiber, J. H., Wijns, W., Brand, M., Kooijman, C. J., Katen, H. J., & Hugenholtz, P. G. (1984). Assessment of percutaneous transluminal coronary angioplasty by quantitative coronary angiography: Diameter versus densitometric area measurements. The American Journal of Cardiology, 54(6), 482–488.
Foley, D. P., Escaned, J., Strauss, B. H., Mario, C., Haase, J., Keane, D., Hermans, W. R., Rensing, B. J., Feyter, P. J., & Serruys, P. W. (1994). Quantitative coronary angiography (qca) in interventional cardiology: Clinical application of qca measurements. Progress in Cardiovascular Diseases, 36(5), 363–384.
Garrone, P., & BIONDI-ZOCCAI, G., Salvetti, I., Sina, N., Sheiban, I., Stella, P.R., & Agostoni, P. (2009). Quantitative coronary angiography in the current era: Principles and applications. Journal of Interventional Cardiology, 22(6), 527–536.
Breininger, K., Albarqouni, S., Kurzendorfer, T., Pfister, M., Kowarschik, M., & Maier, A. (2018). Intraoperative stent segmentation in x-ray fluoroscopy for endovascular aortic repair. International Journal of Computer Assisted Radiology and Surgery, 13, 1221–1231.
Lesage, D., Angelini, E. D., Bloch, I., & Funka-Lea, G. (2009). A review of 3d vessel lumen segmentation techniques: Models, features and extraction schemes. Medical Image Analysis, 13(6), 819–845.
Zhao, F., Chen, Y., Hou, Y., & He, X. (2019). Segmentation of blood vessels using rule-based and machine-learning-based methods: A review. Multimedia Systems, 25, 109–118.
Moccia, S., De Momi, E., El Hadji, S., & Mattos, L. S. (2018). Blood vessel segmentation algorithms-review of methods, datasets and evaluation metrics. Computer Methods and Programs in Biomedicine, 158, 71–91.
Frangi, A.F., Niessen, W.J., Vincken, K.L., & Viergever, M.A. (1998). Multiscale vessel enhancement filtering. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI’98: First International Conference Cambridge, MA, USA, October 11–13, 1998 Proceedings 1, pp. 130–137. Springer.
Jerman, T., Pernuš, F., Likar, B., & Špiclin, Ž. (2016). Enhancement of vascular structures in 3d and 2d angiographic images. IEEE Transactions on Medical Imaging, 35(9), 2107–2118.
Nirmala Devi, S., & Kumaravel, N. (2008). Comparison of active contour models for image segmentation in x-ray coronary angiogram images. Journal of Medical Engineering & Technology, 32(5), 408–418.
Pham, C.-H., Ducournau, A., Fablet, R., & Rousseau, F. (2017). Brain mri super-resolution using deep 3d convolutional networks. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), pp. 197–200. IEEE.
Khokhar, M., Talpur, S., Khowaja, S.A., & Shah, R.A. (2018). A novel curvature feature embedded level set method for image segmentation of coronary angiograms. In: Trends and Advances in Information Systems and Technologies: Volume 2 6, pp. 831–841. Springer.
Brieva, J., Gonzalez, E., Gonzalez, F., Bousse, A., & Bellanger, J. (2006). A level set method for vessel segmentation in coronary angiography. In: 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference, pp. 6348–6351. IEEE.
Qin, B., Jin, M., Hao, D., Lv, Y., Liu, Q., Zhu, Y., Ding, S., Zhao, J., & Fei, B. (2019). Accurate vessel extraction via tensor completion of background layer in x-ray coronary angiograms. Pattern Recognition, 87, 38–54.
Shoujun, Z., Jian, Y., Yongtian, W., & Wufan, C. (2010). Automatic segmentation of coronary angiograms based on fuzzy inferring and probabilistic tracking. Biomedical Engineering, 9(1), 1–21.
Manniesing, R., Viergever, M. A., & Niessen, W. J. (2007). Vessel axis tracking using topology constrained surface evolution. IEEE Transactions on Medical Imaging, 26(3), 309–316.
Zou, P., Chan, P., & Rockett, P. (2008). A model-based consecutive scanline tracking method for extracting vascular networks from 2-d digital subtraction angiograms. IEEE Transactions on Medical Imaging, 28(2), 241–249.
Chen, Y., Zhang, Y., Yang, J., Cao, Q., Yang, G., Chen, J., Shu, H., Luo, L., Coatrieux, J.-L., & Feng, Q. (2015). Curve-like structure extraction using minimal path propagation with backtracking. IEEE Transactions on Image Processing, 25(2), 988–1003.
M’hiri, F., Duong, L., Desrosiers, C., & Cheriet, M. (2013). Vessel walker: Coronary arteries segmentation using random walks and hessian-based vesselness filter. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, pp. 918–921. IEEE.
Lv, T., Yang, G., Zhang, Y., Yang, J., Chen, Y., Shu, H., & Luo, L. (2019). Vessel segmentation using centerline constrained level set method. Multimedia Tools and Applications, 78, 17051–17075.
Ma, G., Yang, J., & Zhao, H. (2020). A coronary artery segmentation method based on region growing with variable sector search area. Technology and Health Care, 28(S1), 463–472.
Xia, S., Zhu, H., Liu, X., Gong, M., Huang, X., Xu, L., Zhang, H., & Guo, J. (2019). Vessel segmentation of x-ray coronary angiographic image sequence. IEEE Transactions on Biomedical Engineering, 67(5), 1338–1348.
Taghizadeh Dehkordi, M., Doost Hoseini, A. M., Sadri, S., & Soltanianzadeh, H. (2014). Local feature fitting active contour for segmenting vessels in angiograms. IET Computer Vision, 8(3), 161–170.
Kerkeni, A., Benabdallah, A., Manzanera, A., & Bedoui, M. H. (2016). A coronary artery segmentation method based on multiscale analysis and region growing. Computerized Medical Imaging and Graphics, 48, 49–61.
Cervantes-Sanchez, F., Cruz-Aceves, I., Hernandez-Aguirre, A., Solorio-Meza, S., Cordova-Fraga, T., & Aviña-Cervantes, J. G. (2018). Coronary artery segmentation in x-ray angiograms using gabor filters and differential evolution. Applied Radiation and Isotopes, 138, 18–24.
Cervantes-Sanchez, F., Cruz-Aceves, I., Hernandez-Aguirre, A., Hernandez-Gonzalez, M. A., & Solorio-Meza, S. E. (2019). Automatic segmentation of coronary arteries in x-ray angiograms using multiscale analysis and artificial neural networks. Applied Sciences, 9(24), 5507.
Nasr-Esfahani, E., Karimi, N., Jafari, M. H., Soroushmehr, S. M. R., Samavi, S., Nallamothu, B., & Najarian, K. (2018). Segmentation of vessels in angiograms using convolutional neural networks. Biomedical Signal Processing and Control, 40, 240–251.
Jo, K., Kweon, J., Kim, Y.-H., & Choi, J. (2018). Segmentation of the main vessel of the left anterior descending artery using selective feature mapping in coronary angiography. IEEE Access, 7, 919–930.
Vukicevic, A. M., Çimen, S., Jagic, N., Jovicic, G., Frangi, A. F., & Filipovic, N. (2018). Three-dimensional reconstruction and nurbs-based structured meshing of coronary arteries from the conventional x-ray angiography projection images. Scientific Reports, 8(1), 1711.
Wan, T., Shang, X., Yang, W., Chen, J., Li, D., & Qin, Z. (2018). Automated coronary artery tree segmentation in x-ray angiography using improved hessian based enhancement and statistical region merging. Computer Methods and Programs in Biomedicine, 157, 179–190.
Sameh, S., Azim, M.A., & AbdelRaouf, A. (2017). Narrowed coronary artery detection and classification using angiographic scans. In: 2017 12th International Conference on Computer Engineering and Systems (ICCES), pp. 73–79. IEEE.
Kerkeni, A., Benabdallah, A., & Bedoui, M.H. (2013). Coronary artery multiscale enhancement methods: a comparative study. In: Image Analysis and Recognition: 10th International Conference, ICIAR 2013, Póvoa do Varzim, Portugal, June 26-28, 2013. Proceedings 10, pp. 510–520. Springer.
Truc, P. T., Khan, M. A., Lee, Y.-K., Lee, S., & Kim, T.-S. (2009). Vessel enhancement filter using directional filter bank. Computer Vision and Image Understanding, 113(1), 101–112.
Xu, X., Liu, B., & Zhou, F. (2013). Hessian-based vessel enhancement combined with directional filter banks and vessel similarity. In: 2013 ICME International Conference on Complex Medical Engineering, pp. 80–84. IEEE.
Ma, H., Hoogendoorn, A., Regar, E., Niessen, W. J., & Walsum, T. (2017). Automatic online layer separation for vessel enhancement in x-ray angiograms for percutaneous coronary interventions. Medical Image Analysis, 39, 145–161.
Jin, M., Li, R., Jiang, J., & Qin, B. (2017). Extracting contrast-filled vessels in x-ray angiography by graduated rpca with motion coherency constraint. Pattern Recognition, 63, 653–666.
Sahafi, A., Koulaouzidis, A., & Lalinia, M. (2024). Polypoid lesion segmentation using yolo-v8 network in wireless video capsule endoscopy images. Diagnostics 14(5)
Lalinia, M., & Sahafi, A. (2024). Colorectal polyp detection in colonoscopy images using yolo-v8 network. Signal, Image and Video Processing, 18(3), 2047–2058.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, pp. 234–241. Springer
Ioffe, S., & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning, pp. 448–456. pmlr.
Ulyanov, D., Vedaldi, A., & Lempitsky, V. (2016). Instance normalization: The missing ingredient for fast stylization. arXiv preprint arXiv:1607.08022.
Ba, J.L., Kiros, J.R., & Hinton, G.E. (2016). Layer normalization. arXiv preprint arXiv:1607.06450.
Wu, Y., & He, K. (2018). Group normalization. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 3–19.
He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778.
Tao, X., Dang, H., Zhou, X., Xu, X., & Xiong, D. (2022). A lightweight network for accurate coronary artery segmentation using x-ray angiograms. Frontiers in Public Health, 10, 892418.
Huang, G., Liu, Z., Van Der Maaten, L., & Weinberger, K.Q. (2017). Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708.
Song, A., Xu, L., Wang, L., Wang, B., Yang, X., Xu, B., Yang, B., & Greenwald, S. E. (2022). Automatic coronary artery segmentation of ccta images with an efficient feature-fusion-and-rectification 3d-unet. IEEE Journal of Biomedical and Health Informatics, 26(8), 4044–4055.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V., & Rabinovich, A. (2015). Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1–9 (2015).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna, Z. (2016). Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826.
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., & Yuille, A. L. (2017). Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40(4), 834–848.
Liu, W., Yang, H., Tian, T., Cao, Z., Pan, X., Xu, W., Jin, Y., & Gao, F. (2022). Full-resolution network and dual-threshold iteration for retinal vessel and coronary angiograph segmentation. IEEE Journal of Biomedical and Health Informatics, 26(9), 4623–4634.
Qin, B., Mao, H., Liu, Y., Zhao, J., Lv, Y., Zhu, Y., Ding, S., & Chen, X. (2022). Robust pca unrolling network for super-resolution vessel extraction in x-ray coronary angiography. IEEE Transactions on Medical Imaging, 41(11), 3087–3098.
Wan, T., Chen, J., Zhang, Z., Li, D., & Qin, Z. (2021). Automatic vessel segmentation in x-ray angiogram using spatio-temporal fully-convolutional neural network. Biomedical Signal Processing and Control, 68, 102646.
Hao, D., Ding, S., Qiu, L., Lv, Y., Fei, B., Zhu, Y., & Qin, B. (2020). Sequential vessel segmentation via deep channel attention network. Neural Networks, 128, 172–187.
Silva, J.L., Menezes, M.N., Rodrigues, T., Silva, B., Pinto, F.J., & Oliveira, A.L. (2021). Encoder-decoder architectures for clinically relevant coronary artery segmentation. arXiv preprint arXiv:2106.11447.
Jun, T. J., Kweon, J., Kim, Y.-H., & Kim, D. (2020). T-net: Nested encoder-decoder architecture for the main vessel segmentation in coronary angiography. Neural Networks, 128, 216–233.
Zhu, X., Cheng, Z., Wang, S., Chen, X., & Lu, G. (2021). Coronary angiography image segmentation based on pspnet. Computer Methods and Programs in Biomedicine, 200, 105897.
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., & Yuille, A.L. (2014). Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv preprint arXiv:1412.7062.
Navarro, F., Shit, S., Ezhov, I., Paetzold, J., Gafita, A., Peeken, J.C., Combs, S.E., & Menze, B.H. (2019). Shape-aware complementary-task learning for multi-organ segmentation. In: Machine Learning in Medical Imaging: 10th International Workshop, MLMI 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13, 2019, Proceedings 10, pp. 620–627. Springer
Ma, Y., Hua, Y., Deng, H., Song, T., Wang, H., Xue, Z., Cao, H., Ma, R., & Guan, H. (2021). Self-supervised vessel segmentation via adversarial learning. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 7536–7545
Park, J., Kweon, J., Kim, Y.I., Back, I., Chae, J., Roh, J.-H., Kang, D.-Y., Lee, P.H., Ahn, J.-M., & Kang, S.-J., et al. (2023). Selective ensemble methods for deep learning segmentation of major vessels in invasive coronary angiography. Medical Physics
Zhang, J., Wang, G., Xie, H., Zhang, S., Huang, N., Zhang, S., & Gu, L. (2020). Weakly supervised vessel segmentation in x-ray angiograms by self-paced learning from noisy labels with suggestive annotation. Neurocomputing, 417, 114–127.
Zhang, Y., Gao, Y., Zhou, G., He, J., Xia, J., Peng, G., Lou, X., Zhou, S., Tang, H., & Chen, Y. (2023). Centerline-supervision multi-task learning network for coronary angiography segmentation. Biomedical Signal Processing and Control, 82, 104510.
Han, T., Ai, D., Li, X., Fan, J., Song, H., Wang, Y., & Yang, J. (2023). Coronary artery stenosis detection via proposal-shifted spatial-temporal transformer in x-ray angiography. Computers in Biology and Medicine, 153, 106546.
He, K., Sun, J., & Tang, X. (2012). Guided image filtering. IEEE Transactions on Pattern Analysis and Machine Intelligence, 35(6), 1397–1409.
Bora, D.J. (2018). An ideal approach for medical color image enhancement. In: Advanced Computational and Communication Paradigms: Proceedings of International Conference on ICACCP 2017, Volume 2, pp. 351–361. Springer.
Polesel, A., Ramponi, G., & Mathews, V. J. (2000). Image enhancement via adaptive unsharp masking. IEEE Transactions on Image Processing, 9(3), 505–510.
Park, T., Khang, S., Jeong, H., Koo, K., Lee, J., Shin, J., & Kang, H. C. (2022). Deep learning segmentation in 2d x-ray images and non-rigid registration in multi-modality images of coronary arteries. Diagnostics, 12(4), 778.
Roy, S. S., Hsu, C., Samaran, A., Goyal, R., Pande, A., & Balas, V. E. (2023). Vessels segmentation in angiograms using convolutional neural network: A deep learning based approach. CMES-Computer Modeling in Engineering & Sciences, 136(1), 241–255.
Yang, S., Kweon, J., Roh, J.-H., Lee, J.-H., Kang, H., Park, L.-J., Kim, D. J., Yang, H., Hur, J., Kang, D.-Y., et al. (2019). Deep learning segmentation of major vessels in x-ray coronary angiography. Scientific Reports, 9(1), 16897.
Gao, Z., Wang, L., Soroushmehr, R., Wood, A., Gryak, J., Nallamothu, B., & Najarian, K. (2022). Vessel segmentation for x-ray coronary angiography using ensemble methods with deep learning and filter-based features. BMC Medical Imaging, 22(1), 10.
Mou, L., Zhao, Y., Chen, L., Cheng, J., Gu, Z., Hao, H., Qi, H., Zheng, Y., Frangi, A., & Liu, J. (2019). Cs-net: Channel and spatial attention network for curvilinear structure segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22, pp. 721–730 (2019). Springer.
Gu, Z., Cheng, J., Fu, H., Zhou, K., Hao, H., Zhao, Y., Zhang, T., Gao, S., & Liu, J. (2019). Ce-net: Context encoder network for 2d medical image segmentation. IEEE Transactions on Medical Imaging, 38(10), 2281–2292.
Liang, D., Qiu, J., Wang, L., Yin, X., Xing, J., Yang, Z., Dong, J., & Ma, Z. (2020). Coronary angiography video segmentation method for assisting cardiovascular disease interventional treatment. BMC Medical Imaging, 20, 1–8.
Li, R.-Q., Bian, G.-B., Zhou, X.-H., Xie, X., Ni, Z.-L., & Hou, Z. (2020). Cau-net: A novel convolutional neural network for coronary artery segmentation in digital substraction angiography. In: Neural Information Processing: 27th International Conference, ICONIP 2020, Bangkok, Thailand, November 23–27, 2020, Proceedings, Part I 27, pp. 185–196. Springer.
Iyer, K., Najarian, C. P., Fattah, A. A., Arthurs, C. J., Soroushmehr, S. R., Subban, V., Sankardas, M. A., Nadakuditi, R. R., Nallamothu, B. K., & Figueroa, C. A. (2021). Angionet: A convolutional neural network for vessel segmentation in x-ray angiography. Scientific Reports, 11(1), 18066.
Algarni, M., Al-Rezqi, A., Saeed, F., Alsaeedi, A., & Ghabban, F. (2022). Multi-constraints based deep learning model for automated segmentation and diagnosis of coronary artery disease in x-ray angiographic images. PeerJ Computer Science, 8, 993.
Menezes, M. N., Lourenço-Silva, J., Silva, B., Rodrigues, T., Francisco, A. R. G., Ferreira, P. C., Oliveira, A. L., & Pinto, F. J. (2022). Development of deep learning segmentation models for coronary x-ray angiography: Quality assessment by a new global segmentation score and comparison with human performance. Revista Portuguesa de Cardiologia, 41(12), 1011–1021.
Nobre Menezes, M., Silva, J. L., Silva, B., Rodrigues, T., Guerreiro, C., Guedes, J. P., Santos, M. O., Oliveira, A. L., & Pinto, F. J. (2023). Coronary x-ray angiography segmentation using artificial intelligence: A multicentric validation study of a deep learning model. The International Journal of Cardiovascular Imaging, 39(7), 1385–1396.
Meng, Y., Du, Z., Zhao, C., Dong, M., Pienta, D., Tang, J., & Zhou, W. (2023). Automatic extraction of coronary arteries using deep learning in invasive coronary angiograms. Technology and Health Care (Preprint), 1–15.
Shen, Y., Chen, Z., Tong, J., Jiang, N., & Ning, Y. (2023). Dbcu-net: deep learning approach for segmentation of coronary angiography images. The International Journal of Cardiovascular Imaging, 39(8), 1571–1579.
Zhang, M., Wang, H., Wang, L., Saif, A., & Wassan, S. (2024). Cidn: A context interactive deep network with edge-aware for x-ray angiography images segmentation. Alexandria Engineering Journal, 87, 201–212.
Funding
Open access funding provided by University of Southern Denmark. This research received no external funding.
Author information
Authors and Affiliations
Contributions
M.L.: Conceptualization, Methodology, Investigation, Visualization, Formal Analysis, Writing Original Draft–Review & Editing. A.S.: Visualization, Formal Analysis, Supervision, Writing–Review & Editing. Both authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no confict of interests.
Consent to participate
Informed consent was obtained from all participants prior to their inclusion in the study.
Consent for publication
We obtained consent from all participants whose data are included in this manuscript.
Ethics approval
The study protocol and all related procedures were reviewed and approved by the Amirkabir University of Technology Review Board.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lalinia, M., Sahafi, A. Coronary Vessel Segmentation in X-ray Angiography Images Using Edge-Based Tracking Method. Sens Imaging 25, 32 (2024). https://doi.org/10.1007/s11220-024-00481-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11220-024-00481-6