Abstract
Long-term trends in fisheries catch are useful to monitor effects of fishing on wild populations. However, fisheries catch data are often aggregated in multi-species complexes, complicating assessments of individual species. Non-target species are often grouped together in this way, but this becomes problematic when increasingly common shifts toward targeting incidental species demand closer management focus at the species level. Species distribution models (SDMs) offer an under-utilised tool to allocate aggregated catch data among species for individual assessments. Here, we present a case study of two shovel-nosed lobsters (Thenus spp.), previously caught incidentally and recorded together in logbook records, to illustrate the design and use of catch allocation SDMs to untangle multi-species data for stock assessments of individual species. We demonstrate how catch allocation SDMs reveal previously masked species-specific catch trends from aggregated data and can identify shifts in fishing behaviour, e.g., changes in target species. Finally, we review key assumptions and limitations of this approach that may arise when applied across a broad geographic or taxonomic scope. Our aim is to provide a template to assist researchers and managers seeking to assess stocks of individual species using aggregated multi-species data.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fisheries logbook data inform assessments of fishing effects on wild populations yet are frequently imperfect for a variety of reasons. A range of approaches have been developed to overcome such shortcomings by building on available data to gap-fill incomplete records. For example, where catch and effort data are lacking from some locations due to spatio-temporal changes in fisheries (e.g., expansion into new areas), missing values can be imputed (Walters 2003; Carruthers et al. 2011). Where fishing location is not documented, records can be associated with target species’ habitats based on catch composition (Stephens and MacCall 2004). Changes in fishing power and gear between early and late time series records can also be accounted for using well established standardisation models (Campbell 2004; Maunder and Punt 2004; Bishop et al. 2008). A less well addressed and increasingly problematic shortcoming in fisheries logbook data is the aggregation of catch data for multiple species into single groups comprising taxonomically similar species. Such aggregated catch data complicate efforts to assess and manage effects of fishing on individual species with different biological and ecological traits (Nakano and Clarke 2006; Saldaña-Ruiz et al. 2017).
Multi-species catch data are often encountered where related or similar species are caught incidentally until shifts in fishing behaviour or conservation status demand closer attention at the species level. Shifts in fisheries target species can occur in response to depletion of previous target stocks or changes in market demand and often prioritise species that can be targeted with existing gear or fishing knowledge to minimise transition costs (Sala et al. 2004; Fulton et al. 2014). Climate change also increasingly drives changes in target species by way of fishery adaptation to species range shifts or changes in species abundance (Pinsky and Fogarty 2012; Rogers et al. 2019; Gillanders et al. 2022). Where species require individual assessments, but logbook data were previously aggregated in multi-species complexes, spatio-temporal differences in species distributions can provide a useful means to allocate aggregated catch data among species.
Species distribution models (SDMs) are a powerful tool for resource and conservation management at regional scales on land (Ferrier et al. 2002; Guisan and Thuiller 2005) and in aquatic systems (Pittman et al. 2007; Moore et al. 2010). Originally, SDMs used relationships between species distributions and abiotic factors like climate, elevation (depth in the marine realm), or habitat structure to predict distributions of taxa in data poor regions (Elith and Leathwick 2009). Advances in SDMs have been closely linked to advances in statistical methods, remote sensing, and computational power, leading to a wide range of SDM applications. Common SDM applications include mapping likely distributions (or potential future distributions) of species of commercial or conservation interest (Ferrier et al. 2002; Maxwell et al. 2009; Pitcher et al. 2012), predicting the spread of invasive species (Peterson 2003; Robertson et al. 2004), and predicting effects of habitat loss or climate change on species distributions (Thomas et al. 2004; Lenoir et al. 2011).
In the fisheries context, the most common application of SDMs involves predicting the distribution of fished species under current or projected climate conditions (Cheung et al. 2009; Brodie et al. 2015; Karp et al. 2023). Studies using SDMs to predict interactions between fishing operations and bycatch or Threatened, Endangered or Protected species (Breen et al. 2016; Catry et al. 2013; Stock et al. 2020) or to model catch per unit effort to develop indices of abundance (Thorson et al. 2020; Hoyle et al. 2024) are also well represented. Other applications of SDMs in fisheries research include investigations of predation on target species (Kempf et al. 2013), prediction of spawning habitats (González-Irusta and Wright 2016), and identification of essential fish habitat for ecosystem-based management (Moore et al. 2016).
A useful but under-utilised application of SDMs in fisheries management is to allocate multi-species catch data to species level. Venables and Dichmont (2004) advocated use of generalised models (GLM, GAM, or GLMM) to allocate aggregated catch records among species based on species distributions. In this way, historical multi-species catch data were allocated to species level for two species of Tiger Prawns: Penaeus semisulcatus (Penaeidae) and P. esculentus (Venables and Dichmont 2004). The need for species-specific assessments has grown due to increased conservation and management focus on individual species since the work of Venables and Dichmont (2004) and is likely to grow further with climate change and growing demand for fisheries resources. Yet despite advances in statistical methods and computing power, use of SDMs to allocate aggregated catch data to species level remains under-utilised in practice.
Here, we present a case study demonstrating the design and decision-making considerations relating to catch allocation models. Catch data for two species of shovel-nosed lobsters (Moreton Bay Bugs; Thenus spp., Scyllaridae) had been recorded together as a multi-species complex in logbooks on the east coast of Queensland, Australia between 1988 and 2021. Moreton Bay Bugs comprise two species: Reef Bugs (Thenus australiensis) and Mud Bugs (T. parindicus), which are distributed around the northern sub-tropical and tropical coast of Australia. Around 80% of landings occur in the Queensland East Coast Otter Trawl Fishery, amounting to ~ 500–700 tonnes per year, with the remainder landed in trawl fisheries operating in other jurisdictions in northern Australia. Aside from the adoption of Turtle Excluder Devices and Bycatch Reduction Devices since the early 2000s, gear used in the fishery has remained broadly similar over the period considered here (1988–2021), with vessels typically deploying two to four demersal trawl nets spread by otter boards and separated by outriggers. Both species were previously caught incidentally but shifts in market demand and fishing effort toward targeted fishing drove a need to assess stocks of both species separately. Aggregated logbook catch data therefore needed to be allocated between species to derive long-term catch rate trends for use as species-specific indices of abundance.
Methods
Model spatial domain and training data
As a starting point, the spatial extent of all available logbook records of Moreton Bay Bug catch was used to inform our model’s spatial domain (Fig. 1A). All available species composition data were then compiled within the model spatial domain to train the model to predict catch composition in data poor areas. Training data were available from: (1) a long-term fishery monitoring survey (n = 1217 sites: O’Neill et al. 2020), (2) a survey of Thenus spp. abundance (n = 103 sites: Louw et al. 2024), (3) a study on biology and behaviour (n = 1 site: Jones 1988), and 4) a study on fishing mortality (n = 147 sites: Courtney 1997) (Fig. 1B). To fill gaps in the training data, a fishery-dependent crew observer program was conducted where fishers photographed their catch to identify species compositions and recorded location and time of data collection (n = 856 sites: McMillan et al. 2023) (Fig. 1B). All surveys used similar otter trawl gear with net mesh sizes from 2″ to 2½” (51–64 mm), i.e., smaller than the minimum legal size of both Thenus species (75 mm carapace width), ensuring catchability was similar among surveys. All surveys were also conducted at similar depths (10–80 m) and at night (6 pm to 6 am). Training data (N = 2324 sites from all sources combined) were intended to inform our response variable, i.e., proportion of Reef Bugs in the catch at each location (with the remainder comprising Mud Bugs).
A The spatial footprint of all Moreton Bay Bug landings from 1988 to 2021 in the Queensland East Coast Otter Trawl Fishery, used to inform our model domain; and B availability of species composition data for Moreton Bay Bugs from all available sources. Sources included two previous studies (Courtney 1997; Jones 1988), a long-term fishery monitoring survey (O’Neill et al. 2020), a survey of Thenus spp. abundance (Louw et al. 2024), and a fishery-dependent crew observer program (McMillan et al. 2023)
Variable selection and habitat data
Both Thenus species prefer similar temperature ranges (Mikami and Greenwood 1997), so a focus on explanatory variables based on habitat preferences was used to model species distributions. Explanatory variables considered potentially useful were sourced from open-source bathymetric and hydrological models (Beaman 2010; Steven et al. 2019), a common approach in marine SDM studies. Species’ habitat preferences can often be leveraged to improve performance of SDMs, but the spatial resolution or coverage of habitat data required by researchers is often unavailable. Increasing access to open-source data repositories and efficient machine learning applications can assist researchers to generate project-specific habitat data to address these issues, something rarely evident in marine SDM studies (Melo-Merino et al. 2020).
Due to known habitat preferences of our candidate species (Mud Bugs prefer finer sediments than Reef Bugs: Jones 1988; Louw et al. 2024), we modelled sediment distributions throughout the study area for use as a habitat variable potentially more informative than depth and hydrology. Sediment modelling was performed along the Queensland east coast from the Torres Strait (10° S) to northern New South Wales (29° S) (Fig. 2A). A spatial domain of 0–200 m was modelled so that habitat layers exceeded the domain modelled for species distributions (5–80 m depth, i.e., the depth range of reported Thenus landings) to avoid edge effects in the SDM. To reduce computational load, the study area was split into four smaller sub-domains of similar size (Fig. 2A). Data from these sub-domains were then mosaicked into a suite of sediment raster layers for the entire model domain.
A Model spatial domain (purple) and regional sub-domains used for habitat modelling (coloured boxes); B availability of sediment point data from the open-source MARS database (blue) and new data collated from various publicly available surveys (maroon; see Table S1)
Extensive sediment point data were sourced from Geoscience Australia’s open-source MARine Sediments (MARS) database (Mathews 2007) and additional sediment data were derived from a range of publicly available studies (Table S1). This resulted in a total of 6761 sites sampled for sediment grain size composition and 5851 sites with calcium carbonate data (Table S1, Fig. 2B). The additional sediment data collated during this project have since been uploaded to the MARS database. A suite of bathymetric (e.g., depth, aspect, slope), hydrologic (e.g., wave and current properties), and reefal explanatory variables (e.g., distance from reef) with potential to influence sediment transport and deposition processes were used to model sediment distributions (Table S2).
Machine learning methods generally outperform other spatial interpolation techniques for the prediction of sediment distribution (Li et al. 2011). Therefore, Random Forest (RF) models were used to produce spatial predictions of sediment habitat properties using the package ‘SPM’ (Spatial Predictive Modelling: Li 2018) in R (v 4.0.5). Random Forest is a machine learning method based on an ensemble of decision trees (Breiman 2001; Kingsford and Salzberg 2008). Advantages of RF are enhanced classification accuracy through the growth of multiple trees, reduced chance of model overfitting due to random subsampling of the dataset to build each tree, and insensitivity to outliers (Breiman 2001).
The cross-validation function in ‘SPM’ (RFCV) was used to determine optimal parameters for all RF models, including testing the maximum number of trees built (ntree, ranging from 500 to 5000 at increments of 500) and the number of variables tried at each node (mtry, ranging from 3 to 9 at increments of 1). Model performance was assessed through ten-fold cross validation (Kohavi 1995). Based on previous studies, the ten-fold cross validation process was repeated for 100 iterations (Li 2013; Li et al. 2013). The error produced by these predictions identified the optimum model using the variance explained by cross validation (VEcv). After model validation, the best ranked model outputs of sediment parameters were used to produce rasters for use as predictor variables in species distribution models (Table S3, Fig. 3).
Example of raster output of sediment properties (sediment mean grain size in this case) modelled throughout the study area for use as predictors of species distributions. Sediment properties were modelled from 0 to 200 m deep to avoid edge effects within the 5–80 m deep species distribution model domain (shown here). Wentworth classifications of sediment type (mud, sand, and gravel) are provided corresponding to mean grain size
Selecting an appropriate response variable
With a suite of potentially influential explanatory variables to model species distributions now available, including habitat parameters, bathymetry, and hydrology (Table S4), an appropriate response variable was developed. Although we are unable to empirically determine whether Mud Bugs and Reef Bugs have differing levels of catchability with commercial otter trawl gear (51–64 mm mesh), we minimised potential differences in catchability by retaining only legal-size individuals (> 75 mm carapace width for both species) for analysis, as these are considered fully recruited to the fishery. This has the added benefit of eliminating any influence of younger life history classes on modelled distributions that may not be relevant to the landed catch.
The intended response variable was the proportion of each species in the legal-size catch at each sampling site. This proportional factor could subsequently be applied to all historical logbook catch records to allocate the catch at each sampling site between species. Trawls resulting in catches of ≤ 1 legal-sized bug were excluded from analyses because sites with single animals cause outlying species habitat preferences when modelling species proportions. Investigation of species composition data both at the scale of individual sampling sites (Fig. 4A) and aggregated to the 0.1˚ logbook reporting grids used in the fishery (Fig. 4B), revealed strong species partitioning, with 89% of sampling sites and 66% of 0.1° logbook reporting grids containing exclusively one species or the other. This type of response variable can be modelled as “proportion of each species” using a zero-and-one-inflated Beta distribution or simplified to “dominant species” (i.e. > 50% of one species or the other) using a binomial regression. We utilised the “dominant species” approach to reduce model complexity, with 0 indicating grids with > 50% catch of Mud Bugs, and 1 indicating grids with > 50% catch of Reef Bugs.
Species dominance (i.e., proportion of the catch comprising Reef Bugs), A by sampling site, and B when sites were aggregated to 0.1° logbook reporting grids. Pink and blue indicate Mud Bug or Reef Bug dominated locations respectively. A, B share a common x-axis. C Number of sampling sites informing each 0.1° grid comprising our training dataset
Model spatial resolution
Selection of an appropriate spatial resolution for modelling will depend on the ecology of candidate species and the spatial footprint of fishing techniques on which logbook data are based. Site-attached species, or species caught using methods with a narrow spatial footprint, e.g., line fishing of reef species with patchy distributions, may require finer spatial resolution of the modelled response than widely dispersed species, or species caught using methods with large spatial footprints (e.g., trawl fishing). The fishery in our case study used 0.1˚ reporting grids to capture the relative spatial ambiguity of trawl catches (i.e., it is not known where exactly along a trawl the catch was distributed). Because of this and the relatively contiguous distribution of our candidate species over large areas, we selected these 0.1° reporting grids as the spatial resolution of our model. Catch composition training data at the scale of individual sampling locations were therefore aggregated at the 0.1° grid scale (Fig. 4C). Mean values for each explanatory variable (habitat parameters, bathymetry, and hydrology) were also calculated at the scale of 0.1˚ grids using the Zonal Statistics tool in ESRI ArcGIS (v 10.8.1). Aggregation of training data at the scale of logbook reporting grids had the added benefits of mitigating spatial autocorrelation arising from uneven density of sampling locations (Fig. 1B) and making model outputs directly relevant to the fishery, such that records from each logbook reporting grid could be attributed to one species or the other.
Model design and application
Modelling of the binomial response “Mud Bug dominant grid” (0) or “Reef Bug dominant grid” (1) was performed using Bernoulli Boosted Regression Trees (BRT) in the ‘gbm’ package in R (Ridgeway 2006), with supporting diagnostics implemented in the ‘dismo’ package (Hijmans et al. 2017). Boosted Regression Trees have several benefits over parametric and semi-parametric models (e.g., GLM, GAM), including their ability to capture complex non-linear relationships, inherently detect and model interactions, iteratively build regression trees from random subsets of the dataset to capture more variance without overfitting, and rank predictor variables by their relative influence (Elith and Leathwick 2009). The number of boosting trees was optimised using the k-fold cross validation capabilities in the ‘dismo’ package (Hijmans et al. 2017).
Although BRTs can be robust to collinearity among predictor variables if the collinearity structure is similar between training and prediction datasets, we opted to conservatively use only sets of variables that were not collinear (r < 0.7: Dormann et al. 2007) in each model build. Seasonality parameters were not included due to the limited movement of Thenus species and the tendency of each species to remain in areas of preferred habitat (Jones 1988; McMillan et al. 2023), making it unlikely that species composition within 0.1˚ grids significantly changes within years. Models were parameterised as follows: learning rate (the contribution or weight of each tree towards the final model) of 0.001, tree complexity (maximum order interactions permitted) of 5, and bag fraction (random subset of the dataset used to build each tree) at the default value of 0.75.
Model outputs were used to assign logbook catch records from 1988 to 2021 to species level based on the predicted dominant species at the location of each catch. Nominal catch per unit effort (CPUE) was then calculated for each species by dividing the annual harvest of each species (in kg) by the annual effort (in hours) recorded in the fishery. Subsequently, a formal stock assessment was undertaken separate to this study, during which CPUE was comprehensively modelled as an index of abundance for stock assessment (Wickens et al. 2023). This process applied a standardisation approach to nominal CPUE that accounted for factors likely to affect catch rates, e.g., changes in fishing power and targeting behaviour (Wickens et al. 2023).
Results
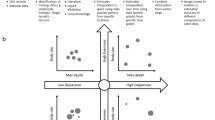
The top ranked model for predicting species distributions included six explanatory variables (by order of relative influence): sediment mean grain size (41.6%), depth (21.8%), sediment medium sand content (21.5%), very fine sand content (6.2%), fine sand content (4.5%), and distance from the coast (4.4%) (Fig. 5A, Table 1). This model accounted for high levels of variance in the dataset (R2 = 0.93) and demonstrated high accuracy at predicting both Mud Bug and Reef Bug species dominance in the training dataset (100% and 99.7% respectively), while also minimising the number of predictors used. Fitted functions for predictor variables indicated Reef Bug dominance in grids characterised by coarse mean grain sizes (low values of Phi), greater depth, distance from coast, and content of medium and fine sands, and lower content of very fine sand (Fig. 5A). Species dominance was also influenced by a significant interaction between mean grain size and depth, whereby Reef Bugs dominated locations at greater depth and with coarse sediment (low Phi), whereas Mud Bugs dominated only locations that were both shallow and with fine sediment (Fig. 5B). Other high-performing models were ranked lower due to declines in variance explained (R2), declines in species-specific classification accuracy, uneven species-specific classification accuracy, and larger numbers of predictors (Table 1).
A Fitted functions for SDM predictor variables. Mean grain size = Phi (1–5 φ = coarse sand to mud), Depth = m, sand fractions = % of total sediment profile, coast distance = degrees longitude. Fitted functions are centred by subtracting their mean. Higher fitted values indicate Reef Bug dominance. Relative influence of each predictor in the model is given in parentheses. All panels share common y-axes. B Interaction plot showing the influence of mean grain size and depth on species distributions. 0 = Mud Bug dominance, 1 = Reef Bug dominance
Across the fishery (N = 1230 grids), most grids were predicted to be dominated by Reef Bugs (861 grids), mirroring the observed dominance of Reef Bugs in the training dataset and field observations reported by fishers. Mud Bugs were predicted to be dominant in shallower inshore waters with fine sediments, particularly in Far North Queensland where these habitats are widely available (Fig. 6).
Modelled outputs for dominant Moreton Bay Bug species in each logbook reporting grid with Thenus catch records. Blue indicates Reef Bug dominant grids; maroon indicates Mud Bug dominant grids. Opaque grids indicate grids for which species dominance observations were available; translucent grids indicate grids for which species dominance was predicted by the best Boosted Regression Tree model
Using the outputs from our SDM, fisheries logbook data were subsequently allocated to species level based on the dominant species in each logbook reporting grid where catches were reported. In cases where candidate species display less distinct partitioning and a proportional response is used, model outputs would allocate a proportion of the total catch at each location to candidate species. In our case study, the allocation of logbook data to species level revealed important information about the composition of the fishery for the first time, indicating that the proportion of the total catch comprising Reef Bugs had increased from 67% at the beginning of logbook records in 1988 to 93% in 2021 (Fig. 7A). Comparison of nominal CPUE for each species revealed that CPUE of Reef Bugs increased markedly from the early 2000s (Fig. 7B).
Discussion
In this study, we have demonstrated the application of a machine learning SDM approach to allocate catch data to species level for assessments where species were previously aggregated in fisheries logbooks. This approach leverages habitat preferences of candidate species to determine likely species compositions at harvest locations. Most input variables used to build SDMs for catch allocation are increasingly available and open access, providing researchers a wide range of opportunities to apply this approach across various systems. Additionally, we illustrated how more complex project-specific habitat data can be generated by researchers using open access explanatory data and machine learning models. We demonstrated this combined approach by modelling sediment properties and the distribution of Moreton Bay Bug species over a large geographic range encompassing the east coast of Queensland, Australia. Due to the strong habitat partitioning observed between our candidate species, we fitted a binomial response achieving almost 100% accuracy in predicting species dominance at 398 locations in the training dataset. By allocating aggregated catch data to species level, our model outputs revealed previously masked harvest trends in the fishery including a marked increase in the proportion of Reef Bugs comprising landings and allowed calculation of nominal CPUE time series for each species. The production of these nominal catch rates facilitated a formal stock assessment, conducted subsequently by Wickens et al. (2023), during which a rigorous standardisation process produced reliable indices of abundance for each species and revealed a pronounced shift to targeting of the larger, more valuable Reef Bugs from the early 2000s. A discussion of key assumptions and limitations that should be considered when applying this approach follows.
Availability of species composition data
To train models for species allocation of aggregated catch data, sufficient information on species distributions is required. These data may be available in the form of survey data, often collected for other purposes, throughout at least part of the candidate species’ distributions. Training data should include information on count or weight of candidate species suitable to derive species’ relative proportions at each site, which can be used as the response variable. In this study, all training data were derived from surveys using similar otter trawl gear likely conferring similar catchability of each species; however, if training data are derived from a diverse range of sources where catchability may vary, survey type should be included as a model term to account for this variation (Nephin et al. 2023).
A cost-effective way of obtaining training data used in our study was a crew observer program, whereby fishers collected photographic information on species compositions at locations they fished. Success of crew observer programs will benefit from data requests and materials being designed to be as efficient and unintrusive as possible to promote uptake by fishers alongside their normal work. In-person visits to distribute and retrieve data collection materials and discuss project aims also help build relationships with fishers and enhance fisher involvement, increasing the available data pool.
Spatio-temporal differences in species distributions improve model performance
To successfully allocate aggregated catch data among species based on species distributions, it is critical that species distributions differ from each other in some aspect of space or time. Evidence of habitat partitioning among species or limited spatial or temporal overlap in distributions will likely improve model performance. In our case study, the candidate species displayed strong preferences for different habitats (sediment grain size and depth) resulting in little spatial overlap. In species with spatial overlap, temporal model terms (e.g., month or season) may identify trends of seasonal abundance in candidate species, as was the case with Tiger Prawns assessed by Venables and Dichmont (2004), where one species was migratory and the other more resident, resulting in seasonal fluctuations in relative abundance.
Incorporating movement and behaviour into SDM design
Careful attention should be paid to aspects of the ecology and biology of candidate species to ensure these are appropriately captured by model design. Modelling distributions of highly mobile species may be complicated by dispersal (Robinson et al. 2011; Saupe et al. 2012; Fabri‐Ruiz et al. 2019). Animal movements are often driven by predictable behaviours like reproductive cycles and/or environmental conditions. Therefore, trialling appropriate temporal model terms will likely be important when dealing with highly mobile and/or seasonally abundant species (Mannocci et al. 2017; Fernandez et al. 2018). Ontogenetic habitat shifts should also be considered when designing SDMs (Robinson et al. 2011; Lloret-Lloret et al. 2020). However, catch allocation SDMs can address this by only using species composition training data from mature animals exceeding the minimum legal size to avoid potentially erroneous habitat associations with immature animals whose distribution may not reflect the landed catch.
Aggregating behaviour can cause clustering that may result in spatial autocorrelation and model overfitting (Dormann et al. 2007). In our case study, dispersion indices and small-scale movements indicated that aggregations associated with mating, spawning, or feeding events were unlikely (Jones 1988). However, in species where aggregating events need to be accounted for, temporally comprehensive training data may capture such trends and appropriate temporal model terms should be trialled to account for these effects. If the focal fishery has temporal closures to avoid harvest of pre-spawning or spawning individuals, it may be unnecessary to account for aggregating events.
Feeding or competitive behaviours will often not affect catch allocation SDMs. Most fisheries species are likely to prefer similar environmental conditions to their prey given their ectothermic physiological requirements. However, in species that can maintain body temperatures above the surrounding environment, e.g. tuna, prey distributions may influence their distributions and should be accounted for as model terms (Robinson et al. 2011). Because fisheries species are typically broadly distributed and are unlikely to attain carrying capacity, where competitive exclusion is most likely to shape population structuring, we consider competition unlikely to affect the composition of fisheries catches. A comprehensive SDM design with appropriate habitat parameters may also adequately describe the distribution of co-distributed competitors, making inclusion of competitor distributions as model terms redundant.
Population status considerations
Processes of habitat selection can result in range expansions into secondary habitats as stocks grow, while declining stocks may contract into smaller core areas around primary preferred habitats (MacCall 1990; Simpson and Walsh 2004; Morfin et al. 2012). Range extensions and contractions may therefore need to be accounted for in cases where stock levels are known to have varied substantially throughout the time series of available records. Ensuring that the model spatial domain captures all areas where catch has been reported throughout the logbook time series should ameliorate any effects of habitat expansion or contraction due to changes in population size.
Sources of explanatory data to model species distributions
A large range of spatio-temporal variables have been used to model marine species distributions, with the selection of appropriate explanatory variables depending on the physiological and ecological requirements of candidate species. Abiotic drivers of species distributions like depth, hydrology, or distance-based metrics are frequently employed. These data are widely available from open-source repositories. Habitat data are less frequently used and rarely generated by researchers themselves (Melo-Merino et al. 2020) but can be valuable predictors of species distributions. This is particularly true for species with strong habitat preferences as in our case study, where sediment characteristics were the most influential drivers of species distributions. Temperature should be used as a model term if candidate species display different temperature preferences.
Distributions of benthic and demersal species are often influenced by seafloor properties due to strong preferences for certain types of habitats (Gray 1974; Auster and Langton 1999; Kostylev et al. 2001). These traits are conducive to SDM approaches. Availability of habitat data from direct habitat surveys is often limited in spatial extent due to logistical expense and complexity. Alternative sources of habitat data at broader spatial scales include open-source geological surveys (as used in our case study) and airborne or satellite photographic surveys (e.g., mapping reefs, seagrass, or kelp beds). Increasing access to multi-beam sonar and underwater video technology will provide detailed high resolution benthic habitat data to inform SDMs (Monk et al. 2012; Courtney et al. 2021).
Pelagic species, due to their three-dimensional use of the water column, may have less connection to physical habitat than benthic taxa. Many pelagic fisheries species also move large distances in response to climatic variation and productivity hotspots such as upwelling fronts or nutrient plumes. Remote sensing data are therefore frequently used to predict distributions of pelagic species (Zagaglia et al. 2004; Lopez et al. 2017; Erauskin‐Extramiana et al. 2019). Sea surface temperature, chlorophyll-a concentration, sea surface height anomaly, wind, climate oscillation indices, and current strength or direction are examples of remotely sensed variables used to model pelagic species distributions.
Ecological relationships as predictors of species distributions
Relationships with co-located taxa may be informative predictors of species distributions, e.g., where taxa are routinely caught together, the presence of well-recorded species may be useful for modelling distributions of species with sparser catch records. In our case study, Tiger Prawn CPUE was positively correlated with Mud Bug CPUE and a useful predictor of Mud Bug distributions at local scales. However, this relationship lost its predictive power at the scale of the entire fishery, highlighting the importance of trialling and selecting model terms at appropriate spatial scales.
Addressing edge effects
Machine learning approaches are sensitive to edge effects, whereby predictions may be less robust when based on spatial extrapolation toward the outer range of the training dataset (Stock et al. 2020). To overcome this limitation, where possible input data should be incorporated into the model framework beyond the edges of the area for which SDM predictions will be made, in effect pushing the spatial edge of reliable input data beyond the edge of the SDM domain. In our case study, explanatory datasets were used to build an input data surface extending beyond the spatial extent of logbook catch records for our candidate species (0–200 m depth) and were subsequently trimmed to the 5–80 m depth range where Moreton Bay Bug species are encountered.
Conclusions
Fisheries catch data reported as multi-species complexes complicate assessments and management at the species level. Assessments of formerly incidental species are increasingly necessary, often requiring untangling of multi-species records to produce species-specific harvest trends. The need for such assessments is growing in response to increasingly common shifts in target species caused by depletion of previous target stocks or range shifts associated with climate change. Species distribution models offer an under-utilised tool for fisheries researchers to allocate catch records among species to inform species-specific assessments. Advances in machine learning and the availability of open-source data platforms provide the opportunity to enhance SDM approaches to allocate aggregated catch data to species level, as well as for researchers to generate their own habitat data to improve model performance.
Data availability
The code and data generated and analysed in this study are available in the following repository: https://doi.org/https://doi.org/10.6084/m9.figshare.25458676.
References
Auster PJ, Langton RW (1999) The effects of fishing on fish habitat. Paper presented at the American Fisheries Society Symposium. 22:150–187
Beaman R (2010) Project 3DGBR: a high-resolution depth model for the Great Barrier Reef and Coral Sea. Marine and Tropical Sciences Research Facility Project 25i1a Final Report: 13
Bishop J, Venables W, Dichmont C, Sterling D (2008) Standardizing catch rates: Is logbook information by itself enough? ICES J Mar Sci 65:255–266. https://doi.org/10.1093/icesjms/fsm179
Breen P, Brown S, Reid D, Rogan E (2016) Modelling cetacean distribution and mapping overlap with fisheries in the northeast Atlantic. Ocean Coast Manag 134:140–149. https://doi.org/10.1016/j.ocecoaman.2016.09.004
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Brodie S, Hobday AJ, Smith JA, Everett JD, Taylor MD, Gray CA, Suthers IM (2015) Modelling the oceanic habitats of two pelagic species using recreational fisheries data. Fish Oceanogr 24:463–477. https://doi.org/10.1111/fog.12122
Campbell RA (2004) CPUE standardisation and the construction of indices of stock abundance in a spatially varying fishery using general linear models. Fish Res 70:209–227. https://doi.org/10.1016/j.fishres.2004.08.026
Carruthers TR, Ahrens RN, McAllister MK, Walters CJ (2011) Integrating imputation and standardization of catch rate data in the calculation of relative abundance indices. Fish Res 109:157–167. https://doi.org/10.1016/j.fishres.2011.01.033
Catry P, Lemos R, Brickle P, Phillips RA, Matias R, Granadeiro JP (2013) Predicting the distribution of a threatened albatross: the importance of competition, fisheries and annual variability. Prog Oceanogr 110:1–10. https://doi.org/10.1016/j.pocean.2013.01.005
Cheung WW, Lam VW, Sarmiento JL, Kearney K, Watson R, Pauly D (2009) Projecting global marine biodiversity impacts under climate change scenarios. Fish Fish 10:235–251. https://doi.org/10.1111/j.1467-2979.2008.00315.x
Courtney AJ, Daniell J, French S, Leigh GM, Yang WH, Campbell MJ et al (2021) Improving mortality rate estimates for management of the Queensland saucer scallop fishery. Fisheries Research and Development Corporation (FRDC) Final Report Project No. 2017/048, p 259
Courtney AJ (1997) A study of the biological parameters associated with yield optimisation of Moreton Bay bugs, Thenus spp. Fisheries Research and Development Corporation (FRDC) Final Report Project No. 92/102, p 45
Dormann CF, McPherson JM, Araújo MB, Bivand R, Bolliger J, Carl G et al (2007) Methods to account for spatial autocorrelation in the analysis of species distributional data: a review. Ecography 30:609–628. https://doi.org/10.1111/j.2007.0906-7590.05171.x
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Ann Rev Ecol Evol Syst 40:677–697. https://doi.org/10.1146/annurev.ecolsys.110308.120159
Erauskin-Extramiana M, Arrizabalaga H, Hobday AJ, Cabré A, Ibaibarriaga L, Arregui I et al (2019) Large-scale distribution of tuna species in a warming ocean. Glob Chang Biol 25:2043–2060. https://doi.org/10.1111/gcb.14630
Fabri-Ruiz S, Danis B, David B, Saucède T (2019) Can we generate robust species distribution models at the scale of the Southern Ocean? Divers Distrib 25:21–37. https://doi.org/10.1038/s41598-020-73262-2
Fernandez M, Yesson C, Gannier A, Miller P, Azevedo J (2018) A matter of timing: how temporal scale selection influences cetacean ecological niche modelling. Mar Ecol Prog Ser 595:217–231. https://doi.org/10.3354/meps12551
Ferrier S, Watson G, Pearce J, Drielsma M (2002) Extended statistical approaches to modelling spatial pattern in biodiversity in northeast New South Wales. I Spec Level Modell Biodivers Conserv 11:2275–2307. https://doi.org/10.1023/A:1021302930424
Fulton EA, Smith AD, Smith DC, Johnson P (2014) An integrated approach is needed for ecosystem based fisheries management: insights from ecosystem-level management strategy evaluation. PLoS ONE 9:e84242. https://doi.org/10.1371/journal.pone.0084242
Gillanders BM, McMillan MN, Reis-Santos P, Baumgartner LJ, Brown LR, Conallin J et al (2022) Climate change and fishes in estuaries. In: Whitfield AK, Able KW, Stephen JM, Elliott M (eds) Fish and fisheries in Estuaries: a global perspective. Wiley, New York. https://doi.org/10.1002/9781119705345.ch7
González-Irusta J, Wright P (2016) Spawning grounds of haddock (Melanogrammus aeglefinus) in the North Sea and West of Scotland. Fish Res 183:180–191. https://doi.org/10.1016/j.fishres.2016.05.028
Gray JS (1974) Animal-sediment relationships. Oceanogr Mar Biol Ann Rev 12:223–261
Guisan A, Thuiller W (2005) Predicting species distribution: offering more than simple habitat models. Ecol Lett 8:993–1009. https://doi.org/10.1111/j.1461-0248.2005.00792.x
Hijmans RJ, Phillips S, Leathwick J, Elith J, Hijmans MRJ (2017) Package ‘dismo.’ Circles 9:1–68
Hoyle SD, Campbell RA, Ducharme-Barth ND, Grüss A, Moore BR, Thorson JT et al (2024) Catch per unit effort modelling for stock assessment: a summary of good practices. Fish Res 269:106860. https://doi.org/10.1016/j.fishres.2023.106860
Jones CM (1988) The biology and behaviour of bay lobsters, Thenus spp.(Decapoda: Scyllaridae), in northern Queensland, Australia. Ph.D. Thesis, University of Queensland
Karp MA, Brodie S, Smith JA, Richerson K, Selden RL, Liu OR et al (2023) Projecting species distributions using fishery-dependent data. Fish Fish 24:71–92. https://doi.org/10.1111/faf.12711
Kempf A, Stelzenmüller V, Akimova A, Floeter J (2013) Spatial assessment of predator–prey relationships in the North Sea: the influence of abiotic habitat properties on the spatial overlap between 0-group cod and grey gurnard. Fish Oceanogr 22:174–192. https://doi.org/10.1111/fog.12013
Kingsford C, Salzberg SL (2008) What are decision trees? Nat Biotechnol 26:1011–1013. https://doi.org/10.1038/nbt0908-1011
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. Paper presented at: international joint conference on artificial intelligence, August 20–25 1995, Montreal, Canada
Kostylev VE, Todd BJ, Fader GB, Courtney R, Cameron GD, Pickrill RA (2001) Benthic habitat mapping on the Scotian Shelf based on multibeam bathymetry, surficial geology and sea floor photographs. Mar Ecol Prog Ser 219:121–137. https://doi.org/10.3354/meps219121
Lenoir S, Beaugrand G, Lecuyer E (2011) Modelled spatial distribution of marine fish and projected modifications in the North Atlantic Ocean. Glob Chang Biol 17:115–129. https://doi.org/10.1111/j.1365-2486.2010.02229.x
Li J (2018) A new R package for spatial predictive modelling. SPM, Brisbane
Li J, Heap AD, Potter A, Daniell JJ (2011) Application of machine learning methods to spatial interpolation of environmental variables. Environ Model Softw 26:1647–1659. https://doi.org/10.1016/j.envsoft.2011.07.004
Li J, Siwabessy J, Tran M, Huang Z, Heap A (2013) Predicting seabed hardness using random forest in R. In: Zhao Y, Cen Y (eds) Data mining applications with R. Academic Press, Cambridge
Li J (2013) Predictive modelling using random forest and its hybrid methods with geostatistical techniques in marine environmental geosciences. Paper presented at the The proceedings of the Eleventh Australasian Data Mining Conference (AusDM 2013), Canberra, Australia
Lloret-Lloret E, Navarro J, Giménez J, López N, Albo-Puigserver M, Pennino MG, Coll M (2020) The seasonal distribution of a highly commercial fish is related to ontogenetic changes in its feeding strategy. Front Mar Sci 7:566686. https://doi.org/10.3389/fmars.2020.566686
Lopez J, Moreno G, Lennert-Cody C, Maunder M, Sancristobal I, Caballero A, Dagorn L (2017) Environmental preferences of tuna and non-tuna species associated with drifting fish aggregating devices (DFADs) in the Atlantic Ocean, ascertained through fishers’ echo-sounder buoys. Deep Sea Res Part II Top Stud Oceanogr 140:127–138. https://doi.org/10.1016/j.dsr2.2017.02.007
Louw N, McMillan MN, Daniell J, Gardiner N, Roberts E (2024) Habitat partitioning in Moreton Bay Bug species to inform fisheries management. Fish Res 273:106956. https://doi.org/10.1016/j.fishres.2024.106956
MacCall AD (1990) Dynamic geography of marine fish populations. Washington University Press, Washington
Mannocci L, Boustany AM, Roberts JJ, Palacios DM, Dunn DC, Halpin PN et al (2017) Temporal resolutions in species distribution models of highly mobile marine animals: recommendations for ecologists and managers. Divers Distrib 23:1098–1109. https://doi.org/10.1111/ddi.12609
Mathews EJ, Heap AD, Woods M (2007) Inter-reefal seabed sediments and geomorphology of the Great Barrier Reef: a spatial analysis. Geosci Aust Rec 2007(09):140
Maunder MN, Punt AE (2004) Standardizing catch and effort data: a review of recent approaches. Fish Res 70:141–159. https://doi.org/10.1016/j.fishres.2004.08.002
Maxwell D, Stelzenmüller V, Eastwood P, Rogers S (2009) Modelling the spatial distribution of plaice (Pleuronectes platessa), sole (Solea solea) and thornback ray (Raja clavata) in UK waters for marine management and planning. J Sea Res 61:258–267. https://doi.org/10.1016/j.seares.2008.11.008
McMillan MN, Leahy SM, Daniell JJ, Louw N, Roberts EM, Wickens M et al (2023) Determining the spatial distribution and abundance indices for Moreton Bay Bugs, Thenus parindicus and Thenus australiensis in Queensland to improve stock assessment and management. Fisheries Research and Development Corporation (FRDC) Final Report Project No. 2020/020
Melo-Merino SM, Reyes-Bonilla H, Lira-Noriega A (2020) Ecological niche models and species distribution models in marine environments: a literature review and spatial analysis of evidence. Ecol Model 415:108837. https://doi.org/10.1016/j.ecolmodel.2019.108837
Mikami S, Greenwood JG (1997) Complete development and comparative morphology of larval Thenus orientalis and Thenus Sp. (Decapoda: Scyllaridae) reared in the laboratory. J Crustac Biol 17:289–308. https://doi.org/10.1163/193724097X00332
Monk J, Ierodiaconou D, Harvey E, Rattray A, Versace VL (2012) Are we predicting the actual or apparent distribution of temperate marine fishes? PLoS ONE 7:e34558. https://doi.org/10.1371/journal.pone.0034558
Moore CH, Harvey ES, Van Niel K (2010) The application of predicted habitat models to investigate the spatial ecology of demersal fish assemblages. Mar Biol 157:2717–2729. https://doi.org/10.1007/s00227-010-1531-4
Moore CH, Drazen JC, Radford BT, Kelley C, Newman SJ (2016) Improving essential fish habitat designation to support sustainable ecosystem-based fisheries management. Mar Pol 69:32–41. https://doi.org/10.1016/j.marpol.2016.03.021
Morfin M, Fromentin JM, Jadaud A, Bez N (2012) Spatio-temporal patterns of key exploited marine species in the Northwestern Mediterranean Sea. PLoS ONE 7:e37907. https://doi.org/10.1371/journal.pone.0037907
Nakano H, Clarke S (2006) Filtering method for obtaining stock indices by shark species from species-combined logbook data in tuna longline fisheries. Fish Sci 72:322–332. https://doi.org/10.1111/j.1444-2906.2006.01153.x
Nephin J, Thompson PL, Anderson SC, Park AE, Rooper CN, Aulthouse B, Watson J (2023) Integrating disparate survey data in species distribution models demonstrate the need for robust model evaluation. Can J Fish Aquat Sci 80:1869–1889. https://doi.org/10.1139/cjfas-2022-0279
O’Neill MF, Yang W, Wortmann J, Courtney AJ, Leigh GM, Campbell MJ, Filar J (2020) Stock predictions and population indicators for Australia’s east coast saucer scallop fishery. Fisheries Research and Development Corporation (FRDC) Final Report Project No. 2017/057, p 114
Palacios DM, Baumgartner MF, Laidre KL, Gregr EJ (2013) Beyond correlation: integrating environmentally and behaviourally mediated processes in models of marine mammal distributions. Endanger Spec Res 22:191–203. https://doi.org/10.3354/esr00558
Peterson AT (2003) Predicting the geography of species’ invasions via ecological niche modeling. Q Rev Biol 78:419–433. https://doi.org/10.1086/378926
Pinsky ML, Fogarty M (2012) Lagged social-ecological responses to climate and range shifts in fisheries. Clim Change 115:883–891. https://doi.org/10.1007/s10584-012-0599-x
Pitcher CR, Lawton P, Ellis N, Smith SJ, Incze LS, Wei CL et al (2012) Exploring the role of environmental variables in shaping patterns of seabed biodiversity composition in regional-scale ecosystems. J Appl Ecol 49:670–679. https://doi.org/10.1111/j.1365-2664.2012.02148.x
Pittman S, Christensen J, Caldow C, Menza C, Monaco M (2007) Predictive mapping of fish species richness across shallow-water seascapes in the Caribbean. Ecol Model 204:921. https://doi.org/10.1016/j.ecolmodel.2006.12.017
Ridgeway G (2006) Gbm: generalized boosted regression models. R Package Vers 1(3):55
Robertson MP, Villet MH, Palmer AR (2004) A fuzzy classification technique for predicting species’ distributions: applications using invasive alien plants and indigenous insects. Divers Distrib 10:461–474. https://doi.org/10.1111/j.1366-9516.2004.00108.x
Robinson L, Elith J, Hobday A, Pearson R, Kendall B, Possingham H, Richardson A (2011) Pushing the limits in marine species distribution modelling: lessons from the land present challenges and opportunities. Glob Ecol Biogeogr 20:789–802. https://doi.org/10.1111/j.1466-8238.2010.00636.x
Rogers LA, Griffin R, Young T, Fuller E, St. Martin K, Pinsky ML (2019) Shifting habitats expose fishing communities to risk under climate change. Nat Clim Change 9:512–516. https://doi.org/10.1038/s41558-019-0503-zz
Sala E, Aburto-Oropeza O, Reza M, Paredes G, López-Lemus LG (2004) Fishing down coastal food webs in the Gulf of California. Fisheries 29:19–25. https://doi.org/10.1577/1548-8446(2004)29[19:FDCFWI]2.0.CO;2
Saldaña-Ruiz LE, Sosa-Nishizaki O, Cartamil D (2017) Historical reconstruction of Gulf of California shark fishery landings and species composition, 1939–2014, in a data-poor fishery context. Fish Res 195:116–129. https://doi.org/10.1016/j.fishres.2017.07.011
Saupe EE, Barve V, Myers CE, Soberón J, Barve N, Hensz CM et al (2012) Variation in niche and distribution model performance: the need for a priori assessment of key causal factors. Ecol Model 237:11–22. https://doi.org/10.1016/j.ecolmodel.2012.04.001
Simpson MR, Walsh SJ (2004) Changes in the spatial structure of Grand Bank yellowtail flounder: testing MacCall’s basin hypothesis. J Sea Res 51:199–210. https://doi.org/10.1016/j.seares.2003.08.007
Stephens A, MacCall A (2004) A multispecies approach to subsetting logbook data for purposes of estimating CPUE. Fish Res 70:299–310. https://doi.org/10.1016/j.fishres.2004.08.009
Steven AD, Baird ME, Brinkman R, Car NJ, Cox SJ, Herzfeld M et al (2019) eReefs: an operational information system for managing the Great Barrier Reef. J Oper Oceanogr 12:12–28. https://doi.org/10.1080/1755876X.2019.1650589
Stock BC, Ward EJ, Eguchi T, Jannot JE, Thorson JT, Feist BE, Semmens BX (2020) Comparing predictions of fisheries bycatch using multiple spatiotemporal species distribution model frameworks. Can J Fish Aquat Sci 77:146–163. https://doi.org/10.1139/cjfas-2018-0281
Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC et al (2004) Extinction risk from climate change. Nature 427:145–148. https://doi.org/10.1038/nature02121
Thorson JT, Maunder MN, Punt E (2020) The development of spatio-temporal models of fishery catch-per-unit-effort data to derive indices of relative abundance. Fish Res 230:105611. https://doi.org/10.1016/j.fishres.2020.105611
Venables WN, Dichmont CM (2004) A generalised linear model for catch allocation: an example from Australia’s Northern Prawn Fishery. Fish Res 70:409–426. https://doi.org/10.1016/j.fishres.2004.08.017
Walters C (2003) Folly and fantasy in the analysis of spatial catch rate data. Can J Fish Aquat Sci 60:1433–1436. https://doi.org/10.1139/f03-152
Wickens ME, Hillcoat KB, Lovett RA, Fox, AR, McMillan MN (2023) Stock assessment of Moreton Bay Bugs (Thenus australiensis and Thenus parindicus) in Queensland, Australia with data to December 2021. Technical Report. State of Queensland, Brisbane
Zagaglia CR, Lorenzzetti JA, Stech JL (2004) Remote sensing data and longline catches of yellowfin tuna (Thunnus albacares) in the equatorial Atlantic. Remote Sens Environ 93:267–281. https://doi.org/10.1016/j.rse.2004.07.015
Acknowledgements
This project was funded by the Fisheries Research and Development Corporation (Project No. FRDC 2020-022) and the Queensland Department of Agriculture and Fisheries. The authors acknowledge the valuable contribution of species composition data and advice by Tony Courtney and Clive Jones as well as Jason McGilvray and all Fisheries Queensland staff involved in the Saucer Scallop long-term monitoring project. Sincere thanks to all commercial fishers in the East Coast Otter Trawl Fishery that contributed to the crew observer program. We also appreciate the provision of harvest and catch rate data by the stock assessment team at Fisheries Queensland.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no financial or competing interests to declare relevant to this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
McMillan, M.N., Leahy, S.M., Hillcoat, K.B. et al. Untangling multi-species fisheries data with species distribution models. Rev Fish Biol Fisheries (2024). https://doi.org/10.1007/s11160-024-09863-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11160-024-09863-1