Abstract
Key message
Physiological and iTRAQ based proteomic analysis provided new insights into potassium deficiency stress response in alligator weed root.
Abstract
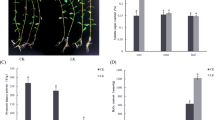
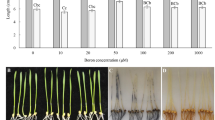
Alligator weed (Alternanthera philoxeroides) has a strong ability to adapt to potassium deficiency (LK) stress. Proteomic changes in response to this stress are largely unknown in alligator weed. In this study, we investigated physiological and molecular mechanisms under LK using isobaric tags for relative and absolute quantitation to characterize proteome-level changes in this plant. First, root physiology, 2, 3, 5-Triphenyl-trazolium chloride (TTC) assay and peroxidase activity were significantly altered after 10 and 15 days of LK treatment. The comparative proteomic analysis suggested a total of 375 proteins were differential abundance proteins. The proteomic results were verified by western blot assays and quantitative real-time PCR. Correlation analysis of transcription and proteomics suggested protein processing in the endoplasmic reticulum, endocytosis, and spliceosome pathways were significantly enriched. The protein responsible for energy metabolism, signal sensing and transduction and protein degradation played crucial roles in this stress. Twelve ubiquitin pathway related proteins were identified in our study, among them 11 proteins were up-regulated. All protein ubiquitination of lysine using pan antibodies were also increased after LK treatment. Our study provide a valuable insights of molecular mechanism underlying LK stress response in alligator weed roots and afford a vital basis to further study potassium nutrition molecular breeding of other plant species.










Similar content being viewed by others
References
Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y (2009) Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol 150:772–785
Bagher R, Bashir H, Ahmad J, Iqbal M, Qureshi MI (2015) Spinach (Spinacia oleracea L.) modulates its proteome differentially in response to salinity, cadmium and their combination stress. Plant Physiol Biochem 97:235e245
Bouthour D, Kalai T, Chaffei HC, Gouia H, Corpas FJ (2015) Differential response of NADP-dehydrogenases and carbon metabolism in leaves and roots of two durum wheat (Triticum durum Desf.) cultivars (Karim and Azizi) with different sensitivities to salt stress. Plant Physiol 179:56–63
Chérei I, Lefoulon C, Boeglin M, Sentenac H (2014) Molecular mechanisms involved in plant adaptation to low K+ availability. J Exp Bot 65:833–848
Cho EK, Choi YJ (2009) A nuclear-localized Hsp70 confers thermoprotective activity and drought-stress tolerance on plants. Biotech Lett 31:597–606
Deng G, Liu LJ, Zhong XY, Lao CY, Wang HY, Wang B (2014) Comparative proteome analysis of the response of ramie under N, P and K deficiency. Planta 239:1175–1186
Galon Y, Aloni R, Nachmias D, Snir O, Feldmesser E, Scrase-Field S (2010) Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232:165–178
Gao L, Geng Y, Li B, Chen J, Yang J (2010) Genome-wide DNA methylation alterations of Alternanthera philoxeroides in natural and manipulated habitats: implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant Cell Environ 33:1820–1827
Hong JP, Takeshi Y, Kondou Y, Schachtman DP, Matsui M, Shin R (2013) Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol 54:1478–71490
Hu C, Lin S, Chi W, Charng Y (2012) Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermos tolerance to chronic heat stress in Arabidopsis. Plant Physiol 158:747–758
Hua X, Wen ZX, Hong MX, Wen WM, Zhen YH, Mi M (2013) The ER luminal binding protein (BiP) alleviates Cd(2+)-induced programmed cell death through endoplasmic reticulum stress-cell death signaling pathway in tobacco cells. J Plant Physiol 170:1434–1441
Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21:607–621
Kang JG, Pyo YJ, Cho JW, Cho MH (2004) Comparative proteome analysis of differentially expressed proteins induced by K+ deficiency in Arabidopsis thaliana. Proteomics 4:3549–3559
Kihara T, Wada T, Suzuki Y, Hara T, Koyama H (2003) Alteration of citrate metabolism in cluster roots of white lupin. Plant Cell Physiol 44:901–908
Kim YH, Bae JM, Huh GH (2010) Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress. Plant Cell Rep 29:779–791
Kim MJ, Ruzicka D, Shin R, Schachtman DP (2012) The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol Plant 5:1042–1057
Koegi M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635–644
Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S (2011) Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol Plant 4:527–536
Latz A, Becker D, Hekman M, Müller T, Beyhl D, Marten I (2007) TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J 52:449–459
Lee MH, Jung C, Lee J, Kim SY, Lee Y, Hwang I (2011) An Abidopsis prenylated rab acceptor 1 isoform, ATPRA1.B6, displays differential inhibitory effects on anterograde trafficking of proteins at the endoplasmic reticulum. Plant Physiol 157:645–658
Li K, Xu C, Li Z, Zhang K, Yang A, Zhang J (2008) Comparative proteome analyses of phosphorus responses in maize (Zea mays L.) roots of wild-type and a low-p-tolerant mutant reveal root characteristics associated with phosphorus efficiency. Plant J Cell Mol Biol 55:927–939
Li N, Zhang Z, Zhang W, Wei Q (2011) Calcineurin b subunit interacts with proteasome subunit alpha type 7 and represses hypoxia-inducible factor-1α activity via the proteasome pathway. BiochemBiophys Res Commun 405:468–472
Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26:3387–3402
Li LQ, Xu L, Wang XY, Pan G, Lu LM (2015) De novo characterization of the alligator weed (Alternanthera philoxeroides L.) transcriptome illuminates gene expression under potassium deprivation. J Genet 94:95–104
Li GZ, Wu YF, Liu GY, Xiao XH, Wang PF, Gao T (2017) Large-scale proteomics combined with transgenic experiments demonstrates an important role of jasmonic acid in potassium deficiency response in wheat and rice. Mol Cell Proteom 16:1889–1905
Lin Z, Lucy A, Rachel H, Don G (2008) LeCTR2, a CTR1-like protein kinase from tomato, plays a role in ethylene signalling, development and defence. Plant J 54:1083–1093
Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280:1943–1945
Luo P, Shen Y, Jin S, Huang S, Cheng X, Wang Z (2016) Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci 245:35–49
Ma QH, Luo HR (2015) Biochemical characterization of caffeoyl coenzyme a 3-o-methyltransferase from wheat. Planta 242:113–122
Maathuis FJ (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12:250–258
Pandey A, Mann M (2000) Proteomics to study genes and genomes. Nature 40:837–846
Ragel P, Ródenas R, García-Martín E, Andrés Z, Villalta I, Nieves-Cordones M (2015) The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol 169:2863–2873
Sivars U, Aivazian D, Pfeffer SR (2003) Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature 425:856–859
Song Z, Su Y (2013) Distinctive potassium-accumulation capability of alligator weed (Alternanthera philoxeroides) links to high-affinity potassium transport facilitated by K+-uptake systems. Weed Sci 61:77–84
Song I, Kim H, Magesh V, Lee K (2018) Ubiquitin c-terminal hydrolase-L1 plays a key role in angiogenesis by regulating hydrogen peroxide generated by NADPH oxidase 4. Biochem Biophys Res Commun 495:1567–1572
Stone SL (2014) The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front Plant Sci 5:1–10
Sung DY, Kim TH, Komives EA, Mendoza-Cózatl DG, Schroeder JI (2009) ARS5 is a component of the 26S proteasome complex, and negatively regulates thiol biosynthesis and arsenic tolerance in Arabidopsis. Plant J Cell Mol Biol 59:802–813
Takizawa M, Goto A, Watanabe Y (2009) The tobacco ubiquitin-activating enzymes NtE1A and NtE1B are induced by tobacco mosaic virus, wounding and stress hormones. Mol Cells 19:228–231
Walker JE (2013) The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans 1817:1–16
Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64:451–476
Wang XC, Li X, Deng X, Han H, Shi W, Li YL (2007) A protein extraction method compatible with proteomic analysis for the euhalophyte Salicornia europaea. Electrophoresis 28:3976–3987
Wang S, Kurepa J, Smalle JA (2009) The Arabidopsis 26S proteasome subunit RPN1a is required for optimal plant growth and stress responses. Plant Cell Physiol 50:721–1725
Wang L, Liu X, Liang M, Tan F, Liang W, Chen Y (2014) Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-term stress. PloS ONE 9:e83141
Wang XC, Li Q, Jin X, Xiao GH, Liu GJ, Liu NJ, Qin YM (2015) Quantitative proteomics and transcriptomics reveal key metabolic processes associated with cotton fiber initiation. J Proteom 114:16–27
Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L (2006) A protein kinase, interacting with two calcineurin b-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125:1347–1360
Xu W, Shi W, Jia L, Liang J, Zhang J (2012) TFT6 and TFT7, two different members of tomato 14-3-3gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ 35:1393–1406
Yamauchi T, Watanabe K, Fukazawa A, Mori H, Abe F, Kawaguchi K (2014) Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J Exp Bot 65:261–273
Yang P, Smalle J, Lee S, Yan N, Emborg TJ, Vierstra RD (2007) Ubiquitin c-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J 51:441–457
Zhang B, Karnik R, Wang YZ, Wallmeroth N, Blatt MR, Grefen C (2015) The Arabidopsis R-SNARE VAMP721 interacts with KAT1 andKC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27:1697–1717
Zhang Z, Chao M, Wang S, Bu J, Tang J, Li F (2016) Proteome quantification of cotton xylem sap suggests the mechanisms of potassium-deficiency-induced changes in plant resistance to environmental stresses. Sci Rep 6:21060–21075
Zhou HP, Lin HX, Chen S, Becker K, Yang YQ, Zhao JF (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26:1166–1182
Acknowledgements
This research was financially supported by a grant from the National Natural Science Foundation (31070244) and the State Key Laboratory of Plant Physiology and Biochemistry (SKLPPBKF1505 and SKLPPBKF1506). We thanked Xuchu Wang from Hainan Normal University for technical support with the western blot work in the present study.
Author information
Authors and Affiliations
Contributions
LL, XW and LL designed the experiments. LL,WZ, QC performed the experiments, SH, SP, YL analyzed the data. LL and LL wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Lq., Liu, L., Zhuo, W. et al. Physiological and quantitative proteomic analyses unraveling potassium deficiency stress response in alligator weed (Alternanthera philoxeroides L.) root. Plant Mol Biol 97, 265–278 (2018). https://doi.org/10.1007/s11103-018-0738-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0738-5