Abstract
Key message
The application of Protein Contact Networks methodology allowed to highlight a novel response of border region between the two domains to substrate binding.
Abstract
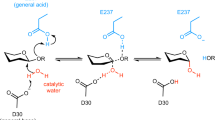
Glycoside hydrolases (GH) are enzymes that mainly hydrolyze the glycosidic bond between two carbohydrates or a carbohydrate and a non-carbohydrate moiety. These enzymes are involved in many fundamental and diverse biological processes in plants. We have focused on the GH32 family, including enzymes very similar in both sequence and structure, each having however clear specificities of substrate preferences and kinetic properties. Structural and topological differences among proteins of the GH32 family have been here identified by means of an emerging approach (Protein Contact network, PCN) based on the formalization of 3D structures as contact networks among amino-acid residues. The PCN approach proved successful in both reconstructing the already known functional domains and in identifying the structural counterpart of the properties of GH32 enzymes, which remain uncertain, like their allosteric character. The main outcome of the study was the discovery of the activation upon binding of the border (cleft) region between the two domains. This reveals the allosteric nature of the enzymatic activity for all the analyzed forms in the GH32 family, a character yet to be highlighted in biochemical studies. Furthermore, we have been able to recognize a topological signature (graph energy) of the different affinity of the enzymes towards small and large substrates.






Similar content being viewed by others
References
Altenbach D, Nüesch E, Meyer AD, Boller T, Wiemken A (2004) The large subunit determines catalytic specificity of barley sucrose:fructan 6-fructosyltransferase and fescue sucrose:sucrose 1-fructosyltransferase. FEBS Lett 567(2–3):214–218. doi:10.1016/j.febslet.2004.04.064
Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S (2004) Network analysis of protein structures identifies functional residues. J Mol Biol 344(4):1135–1146. doi:10.1016/j.jmb.2004.10.055
Bhattacharyya, M, Ghosh S, Vishveshwara S (2016) Protein structure and function: looking through the network of side-chain interactions. Curr Protein Pept Sci 17(1):4–25
Cimini S, Locato V, Vergauwen R, Paradiso A, Cecchini C, Vandenpoel L, Verspreet J et al (2015) Fructan biosynthesis and degradation as part of plant metabolism controlling sugar fluxes during durum wheat kernel maturation. Front Plant Sci 6:89. doi:10.3389/fpls.2015.00089
Cumbo F, Paci P, Santoni D, Di Paola L, Giuliani A. 2014. GIANT: a cytoscape plugin for modular networks. PLoS One. doi:10.1371/journal.pone.0105001
De Ruvo M, Giuliani A, Paci P, Santoni D, Di Paola L (2012) Shedding light on protein–ligand binding by graph theory: the topological nature of allostery. Biophys Chem 165–166:21–29. doi:10.1016/j.bpc.2012.03.001
Ende VW, Coopman M, Clerens S, Vergauwen R, Roy KL, Lammens W, Van Laere A (2011) Unexpected presence of graminan- and levan-type fructans in the evergreen frost-hardy eudicot pachysandra terminalis (Buxaceae): purification, cloning, and functional analysis of a 6-SST/6-SFT enzyme. Plant Physiol 155(1):603–614. doi:10.1104/pp.110.162222
Giuliani, A, Paola LD (2016) Protein as networks: will contact maps hold the promise to represent the ‘structural-formula’ of protein molecules? Curr Protein Pept Sci 17(1):3
Giuliani A, Paola LD, Setola R (2009) Proteins as networks: a mesoscopic approach using haemoglobin molecule as case study. Curr Proteomics 6(4):235–245
Guimerà R, Sales-Pardo M, Amaral LAN (2006) Classes of complex networks defined by role-to-role connectivity profiles. Nat Phys 3:63–69
Gutman I, Zhou B (2006) Laplacian energy of a graph. Linear Algebra Appl 414(1):29–37. doi:10.1016/j.laa.2005.09.008
Henrissat B (1991) A classification of glycosyl hydrolases based sequence similarities amino acid. Biochem J 280(Pt 2):309–316. doi:10.1007/s007920050009
Hothorn M, Van Den Edne W, Lammens W, Rybin V, Scheffzek K (2010) Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc Natl Acad Sci USA. doi:10.1073/pnas.1004481107
Krishnan A, Zbilut JP, Tomita M, Giuliani A (2008) Proteins as networks: usefulness of graph theory in protein science. Curr Protein Pept Sci 9:28–38
Lammens W, Roy KL, Van Laere A, Rabijns A, Van Den Ende W (2008) Crystal structures of arabidopsis thaliana cell-wall invertase mutants in complex with sucrose. J Mol Biol 377(2):378–385. doi:10.1016/j.jmb.2007.12.074
Lammens W, Roy KL, Schroeven L, Van Laere A, Rabijns A, Van Den Ende W (2009) Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J Exp Bot 60(3):727–740. doi:10.1093/jxb/ern333
Lammens W, Roy KL, Yuan S, Vergauwen R, Rabijns A, Van Laere A, Strelkov SV, Van Den Ende W (2012) Crystal structure of 6-SST/6-SFT from pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose. Plant J 70(2):205–219. doi:10.1111/j.1365-313X.2011.04858.x
Le Roy K Verhaest M, Rabijns A, Clerens S, Van Laere A, Van Den Ende W (2007) N-Glycosylation affects substrate specificity of chicory fructan 1-exohydrolase: evidence for the presence of an inulin binding cleft. New Phytol 176:317–324. doi:10.1111/j.1469-8137.2007.02174.x
Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 32:W590–W594. doi:10.1093/nar/gkh477
Matrai J, Lammens W, Jonckheer A, Le Roy K, Rabijns A, Van den Ende W, De Maeyer M (2007) An alternate sucrose binding mode in the e203q arabidopsis invertase mutant: an X-ray crystallography and docking study. Proteins 71:552–564 http://www.rcsb.org/pdb/explore/explore.do?structureId=2OXB
Minic Z (2008) Physiological roles of plant glycoside hydrolases. Planta 723–740. doi:10.1007/s00425-007-0668-y
Oliva G, Di Paola L, Giuliani A, Pascucci F, Setola R (2013) Assessing protein resilience via a complex network approach. In: IEEE 2nd Network Science Workshop (NSW), pp 131–137
Paci P, Di Paola L, Santoni D, De Ruvo M, Giuliani A (2012) Structural and functional analysis of hemoglobin and serum albumin through protein long-range interaction networks. Curr Proteomics 9(3):160–166
Paola DL, De Ruvo M, Paci P, Santoni D, Giuliani A (2013) Protein contact networks: an emerging paradigm in chemistry. Chem Rev 113(3):1598–1613. doi:10.1021/cr3002356
Paola DL, Platania CBiancaM, Oliva G, Setola R, Pascucci F, Giuliani A (2015) Characterization of protein–protein interfaces through a protein contact network approach. Front Bioeng Biotechnol 3:1–10. doi:10.3389/fbioe.2015.00170
Paola D, Luisa GM, Di Venere A, Giuliani A (2016) Exploring the stability of dimers through protein structure topology. Curr Protein Pept Sci 17(1):30–36. doi:10.2174/1389203716666150923104054
Platania CBM, Di Paola L, Leggio GM, Romano GL, Drago F, Salomone S, Bucolo C (2015) Molecular features of interaction between VEGFA and anti-angiogenic drugs used in retinal diseases: a computational approach. Front Pharmacol 6:248
Pons T, Naumoff DG, Martínez-Fleites C, Hernández L (2004) Three acidic residues are at the active site of a beta-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins 54(3):424–432. doi:10.1002/prot.10604
Tasdighian S, Paola LD, De Ruvo M, Paci P, Santoni D, Palumbo P, Mei G, Di Venere A, Giuliani A (2014) Modules identification in protein structures: the topological and geometrical solutions. J Chem Inf Model 54:159–168. doi:10.1021/ci400218v
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. In: Manhold R, Kubinyi H, Timmermann H (eds) Methods and principles in medicinal chemistry, vol 11. Wiley VCH, Weinheim
Trollope KM, van Wyk N, Kotjomela MA, Volschenk H (2015) Sequence and structure-based prediction of fructosyltransferase activity for functional sub-classification of fungal GH32 enzymes. FEBS J. doi:10.1111/febs.13536
Tsai CJ, del Sol A, Nussinov R (2008) Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol 378(1):1–11
Ullman JD, Yannakakis M (1991) High-probability parallel transitive-closure algorithms. SIAM J Comput 20(1):100–125
Verhaest M, Ende WV, Roy KL, De Ranter CJ, Van Laere A, Rabijns A (2005a) X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus. Plant J Cell Mol Biol 41(3):400–411. doi:10.1111/j.1365-313X.2004.02304.x
Verhaest M, Le Roy K, Stefaan S, De Coninck B, Lammens W, De Ranter CJ, Van Laere A, Van Den Ende W, Rabijns A (2005b) Crystallization and preliminary x-ray diffraction study of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr 61:766–768. doi:10.1107/S1744309105021421
Verhaest M, Lammens W, Le Roy K, De Coninck B, De Ranter CJ, Van Laere A, Van Den Ende W, Rabijns A (2006a) X-ray diffraction structure of a cell-wall invertase from Arabidopsis thaliana. Acta Crystallogr 62(Pt 12):1555–1563. doi:10.1107/S0907444906044489
Verhaest M, Lammens W, Le Roy K, De Ranter CJ, Van Laere A, Rabijns A, Van den Ende W (2007) Insights into the fine architecture of the active site of chicory fructan 1-exohydrolase: 1-kestose as substrate vs sucrose as inhibitor. New Phytol 174(1):90–100. doi:10.1111/j.1469-8137.2007.01988.x
Verspreet J, Cimini S (2013) Fructan metabolism in developing wheat (Triticum aestivum L.) kernels. Plant Cell 54(12):2047–2057. doi:10.1093/pcp/pct144
Author contributions
All authors have seen and approved the manuscript and its contents, and that they are aware of the responsibilities connected to authorship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cimini, S., Di Paola, L., Giuliani, A. et al. GH32 family activity: a topological approach through protein contact networks. Plant Mol Biol 92, 401–410 (2016). https://doi.org/10.1007/s11103-016-0515-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0515-2