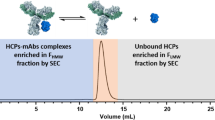
Abstract
Purpose or Objective
Surfactants, including polysorbates and poloxamers, play a crucial role in the formulation of therapeutic proteins by acting as solubilizing and stabilizing agents. They help prevent protein aggregation and adsorption, thereby enhancing the stability of drug substance and products., However, it is important to note that utilizing high concentrations of surfactants in protein formulations can present significant analytical challenges, which can ultimately affect the product characterization.
Methods
In our study, we specifically investigated the impact of elevated surfactant concentrations on the characterization of monoclonal antibodies. We employed various analytical techniques including size-exclusion chromatography (SEC), capillary electrophoresis (CE-SDS), a cell based functional assay, and biophysical characterization.
Results
The findings of our study indicate that higher levels of Polysorbate 80 (PS-80) have adverse effects on the measured purity, biological activity, and biophysical characterization of biologic samples. Specifically, the elevated levels of PS-80 cause analytical interferences, which can significantly impact the accuracy and reliability of analytical studies.
Conclusions
Our study results highlight a significant risk in analytical investigations, especially in studies involving the isolation and characterization of impurities. It is important to be cautious of surfactant concentrations, as they can become more concentrated during common sample manipulations like buffer exchange. Indeed, the research presented in this work emphasizes the necessity to evaluate the impact on analytical assays when there are substantial alternations in the matrix composition. By doing so, valuable insights can be gained regarding potential challenges associated with assay development and characterization of biologics with complex formulations.








Similar content being viewed by others
References
Dimitrov DS. Therapeutic Proteins. In: Voynov V, Caravella JA, editors. Therapeutic Proteins: Methods and Protocols. Totowa, NJ: Humana Press; 2012. p. 1–26.
Mullard A. 2022 FDA approvals. Nat Rev Drug Discov. 2023;22(2):83–8.
Breedveld FC. Therapeutic monoclonal antibodies. The Lancet. 2000;355(9205):735–40.
Waldmann TA. Monoclonal Antibodies in Diagnosis and Therapy. Science. 1991;252(5013):1657–62.
Pinholt C, Hartvig RA, Medlicott NJ, Jorgensen L. The importance of interfaces in protein drug delivery - why is protein adsorption of interest in pharmaceutical formulations? Expert Opin Drug Deliv. 2011;8(7):949–64.
Khan TA, Mahler HC, Kishore RS. Key interactions of surfactants in therapeutic protein formulations: A review. Eur J Pharm Biopharm. 2015;97(Pt A):60–7.
Kim HL, McAuley A, McGuire J. Protein effects on surfactant adsorption suggest the dominant mode of surfactant-mediated stabilization of protein. J Pharm Sci. 2014;103(5):1337–45.
Deechongkit S, Wen J, Narhi LO, Jiang Y, Park SS, Kim J, et al. Physical and biophysical effects of polysorbate 20 and 80 on darbepoetin alfa. J Pharm Sci. 2009;98(9):3200–17.
Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24(10):1241–52.
Chung S, Tian J, Tan Z, Chen J, Lee J, Borys M, et al. Industrial bioprocessing perspectives on managing therapeutic protein charge variant profiles. Biotechnol Bioeng. 2018;115(7):1646–65.
Du Y, Walsh A, Ehrick R, Xu W, May K, Liu H. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. MAbs. 2012;4(5):578–85.
Kahle J, Watzig H. Determination of protein charge variants with (imaged) capillary isoelectric focusing and capillary zone electrophoresis. Electrophoresis. 2018;39(20):2492–511.
Fekete S, Beck A, Veuthey JL, Guillarme D. Ion-exchange chromatography for the characterization of biopharmaceuticals. J Pharm Biomed Anal. 2015;113:43–55.
Neill A, Nowak C, Patel R, Ponniah G, Gonzalez N, Miano D, et al. Characterization of Recombinant Monoclonal Antibody Charge Variants Using OFFGEL Fractionation, Weak Anion Exchange Chromatography, and Mass Spectrometry. Anal Chem. 2015;87(12):6204–11.
Jaag S, Shirokikh M, Lammerhofer M. Charge variant analysis of protein-based biopharmaceuticals using two-dimensional liquid chromatography hyphenated to mass spectrometry. J Chromatogr A. 2021;1636: 461786.
Shi RL, Xiao G, Dillon TM, Ricci MS, Bondarenko PV. Characterization of therapeutic proteins by cation exchange chromatography-mass spectrometry and top-down analysis. MAbs. 2020;12(1):1739825.
Leblanc Y, Ramon C, Bihoreau N, Chevreux G. Charge variants characterization of a monoclonal antibody by ion exchange chromatography coupled on-line to native mass spectrometry: Case study after a long-term storage at +5°C. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;1048:130–9.
Zbacnik TJ, Holcomb RE, Katayama DS, Murphy BM, Payne RW, Coccaro RC, et al. Role of Buffers in Protein Formulations. J Pharm Sci. 2017;106(3):713–33.
Yang K, Hewarathna A, Geerlof-Vidavsky I, Rao VA, Gryniewicz-Ruzicka C, Keire D. Screening of Polysorbate-80 Composition by High Resolution Mass Spectrometry with Rapid H/D Exchange. Anal Chem. 2019;91(22):14649–56.
Chou DK, Krishnamurthy R, Randolph TW, Carpenter JF, Manning MC. Effects of Tween 20 and Tween 80 on the stability of Albutropin during agitation. J Pharm Sci. 2005;94(6):1368–81.
Bartling CM, Andre JC, Howland CA, Hester ME, Cafmeyer JT, Kerr A, et al. Stability Characterization of a Polysorbate 80-Dimethyl Trisulfide Formulation, a Cyanide Antidote Candidate. Drugs R D. 2016;16(1):109–27.
Toutain-Kidd CM, Kadivar SC, Bramante CT, Bobin SA, Zegans ME. Polysorbate 80 inhibition of Pseudomonas aeruginosa biofilm formation and its cleavage by the secreted lipase LipA. Antimicrob Agents Chemother. 2009;53(1):136–45.
Singh SM, Bandi S, Jones DNM, Mallela KMG. Effect of Polysorbate 20 and Polysorbate 80 on the Higher-Order Structure of a Monoclonal Antibody and Its Fab and Fc Fragments Probed Using 2D Nuclear Magnetic Resonance Spectroscopy. J Pharm Sci. 2017;106(12):3486–98.
Lakowicz JR. On spectral relaxation in proteins. Photochem Photobiol. 2000;72(4):421–37.
Randolph TW, Jones LS. Surfactant-protein interactions. Pharm Biotechnol. 2002;13:159–75.
Sinha NJ, Guo R, Misra R, Fagan J, Faraone A, Kloxin CJ, et al. Colloid-like solution behavior of computationally designed coiled coil bundlemers. J Colloid Interface Sci. 2022;606(Pt 2):1974–82.
Gill P, Moghadam TT, Ranjbar B. Differential scanning calorimetry techniques: applications in biology and nanoscience. J Biomol Tech. 2010;21(4):167–93.
Falke S, Betzel C. Dynamic Light Scattering (DLS): principles, perspectives, applications to biological samples. Radiation in Bioanalysis. 2019;8:173–93.
Otzen D. Protein-surfactant interactions: a tale of many states. Biochim Biophys Acta. 2011;1814(5):562–91.
Parkins DA, Lashmar UT. The formulation of biopharmaceutical products. Pharm Sci Technol Today. 2000;3(4):129–37.
Wang W, Wang YJ, Wang DQ. Dual effects of Tween 80 on protein stability. Int J Pharm. 2008;347(1–2):31–8.
Zhou X, Fennema Galparsoro D, Østergaard Madsen A, Vetri V, van de Weert M, Mørck Nielsen H, et al. Polysorbate 80 controls Morphology, structure and stability of human insulin Amyloid-Like spherulites. J Colloid Interface Sci. 2022;606:1928–39.
Horiuchi S, Winter G. CMC determination of nonionic surfactants in protein formulations using ultrasonic resonance technology. Eur J Pharm Biopharm. 2015;92:8–14.
Lutz H, Wilkins R, Carbrello C. Sterile Filtration: Principles, Best Practices and New Developments. In: Kolhe P, Shah M, Rathore N, editors. Sterile Product Development: Formulation, Process, Quality and Regulatory Considerations. New York, NY: Springer New York. 2013 431–59.
Mahler H-C, Huber F, Kishore RSK, Reindl J, Rückert P, Müller R. Adsorption Behavior of a Surfactant and a Monoclonal Antibody to Sterilizing-Grade Filters. J Pharm Sci. 2010;99(6):2620–7.
Acknowledgements
We are thankful to our colleagues Aoshuang Xu, Katie Neal, Jonathan Welch, Kirby Martinez-Fonts for their scientific discussion and help with the experiments.
Funding
The authors would like to acknowledge MRL, Department of Analytical Research & Development (Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA) for research funding to support this work.
Author information
Authors and Affiliations
Contributions
KZ, JS, designed and performed the experiments, analyzed the data and draft the manuscript. PB designed and performed the experiments and analyzed the data. AJP designed the experiments, analyzed the data and refined the manuscript. JRB, RCG and HX provided constructive suggestions and refined the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
All authors declare no conflict of interest related to the work in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, K., Song, J., Bennington, P. et al. Identification of Surfactant Impact on a Monoclonal Antibody Characterization via HPLC-Separation Based and Biophysical Methods. Pharm Res 41, 779–793 (2024). https://doi.org/10.1007/s11095-024-03684-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-024-03684-4