Abstract
Purpose
Normalised prediction distribution errors (npde) are used to graphically and statistically evaluate mixed-effect models for continuous responses. In this study, our aim was to extend npde to time-to-event (TTE) models and evaluate their performance.
Methods
Let V denote a dataset with censored TTE observations. The null hypothesis (H0) is that observations in V can be described by model M. We extended npde to TTE models using imputations to take into account censoring. We then evaluated their performance in terms of type I error and power to detect model misspecifications for TTE data by means of a simulation study with different sample sizes.
Results
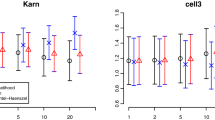
Type I error was found to be close to the expected 5% significance level for all sample sizes tested. The npde were able to detect misspecifications in the baseline hazard as well as in the link between the longitudinal variable and the survival function. The ability to detect model misspecifications increased as the difference in the shape of the survival function became more apparent. As expected, the power also increased as the sample size increased. Imputing the censored events tended to decrease the percentage of rejections.
Conclusions
We have shown that npde can be readily extended to TTE data and that they perform well with an adequate type I error.







Similar content being viewed by others
Abbreviations
- BLQ:
-
Below the limit of quantification
- IIV:
-
Inter-individual variability
- KM:
-
Kaplan Meier
- KMVPC:
-
Kaplan Meier visual predictive check
- NLMEM:
-
Nonlinear mixed-effect models
- npde:
-
Normalised prediction distribution errors
- pd:
-
Prediction discrepancies
- PSA:
-
Prostate-specific antigen
- TTE:
-
Time to event
- VPC:
-
Visual predictive check
References
Holford N. A time to event tutorial for pharmacometricians. CPT Pharmacometrics Syst Pharmacol. 2013;2:e43.
Flynn R. Survival analysis. J Clin Nurs. 2012;21(19–20):2789–97.
Versmissen J, Oosterveer DM, Yazdanpanah M, Defesche JC, Basart DCG, Liem AH, et al. Efficacy of statins in familial hypercholesterolaemia: a long term cohort study. BMJ. 2008;337:a2423.
de Oliveira C, Watt R, Hamer M. Toothbrushing, inflammation, and risk of cardiovascular disease: results from Scottish Health Survey. BMJ. 2010;340:c2451.
Mould D, Upton R. Basic concepts in population modeling, simulation, and model-based drug development. CPT Pharmacomet Syst Pharmacol. 2012;1(9):1–14.
Ibrahim J, Chu H, Chen L. Basic concepts and methods for joint models of longitudinal and survival data. J Clin Oncol. 2010;28(16):2796–801.
Mbogning C, Bleakley K, Lavielle M. Joint modelling of longitudinal and repeated time-to-event data using nonlinear mixed-effects models and the stochastic approximation expectation–maximization algorithm. J Stat Comput Simul. 2015;85(8):1512–28.
Ette E, Williams P. Pharmacometrics: the science of quantitative pharmacology. Hoboken, New Jersey: John Wiley and Sons; 2013.
Food and Drug Administration. Guidance for Industry Population Pharmacokinetics: Center for Drug Evaluation and Research (CDER) and Center for Biologics Evaluation and Research (CBER); 1999. Available from: http://www.fda.gov/downloads/Drugs/Guidances/UCM072137.pdf .
Food and Drug Administration. Guidance for Industry Exposure-Response Relationships– Study Design, Data Analysis, and Regulatory Applications: Center for Drug Evaluation and Research (CDER) and Center for Biologics Evaluation and Research (CBER); 2003. Available from: https://www.fda.gov/ downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm072109.pdf .
Agency EM. Guideline on reporting the results of population pharmacokinetic analysis CHMP; 2007. Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_ guideline/2009/09/WC500003067.pdf .
Marshall S, Burghaus R, Cosson V, Cheung S, Chenel M, DellaPasqua O, et al. Good Practices in Model-Informed Drug Discovery and Development: Practice, Application, and Documentation. CPT Pharmacometrics Syst Pharmacol. 2016;5(3):93–122.
Brendel K, Dartois C, Comets E, Lemmenuel-Diot A, Laveille C, Tranchand B, et al. Are population PK and/or PD models adequately evaluated? A 2002 to 2004 literature survey. Clin Pharmacokinet. 2007;46(3):221–34.
Lavielle M. Mixed effects models for the population approach: models, tasks, methods and tools. London: Chapman and Hall/CRC Biostatistics Series; 2014.
Nguyen T, Mouksassi MS, Holford N, Al-Huniti N, Freedman I, Hooker A, et al. Model evaluation of continuous data pharmacometric models: metrics and graphics. CPT Pharmacometrics Syst Pharmacol. 2017;6(2):87–109.
Beal S, Sheiner L, Boeckmann A, Bauer R. NONMEM Version 7.4. Ellicott City; 1989-2017.
Yano Y, Beal SL, Sheiner LB. Evaluating pharmacokinetic/pharmacodynamic models using the posterior predictive check. J Pharmacokinet Pharmacodyn. 2001;28(2):171–92.
Mentré F, Escolano S. Prediction discrepancies for the evaluation of nonlinear mixed-effects models. J Pharmacokinet Pharmacodyn. 2006;33(3):345–67.
Hooker AC, Staatz CE, Karlsson MO. Conditional weighted residuals (CWRES): a model diagnostic for the FOCE method. Pharm Res. 2007;24(12):2187–97.
Brendel K, Comets E, Laffont C, Laveille C, Mentré F. Metrics for external model evaluation with an application to the population pharmacokinetics of gliclazide. Pharm Res. 2006;23(9):2036–49.
Brendel K, Comets E, Laffont C, Mentré F. Evaluation of different tests based on observations for external model evaluation of population analyses. J Pharmacokinet Pharmacodyn. 2010;37(1):49–65.
Comets E, Brendel K, Mentré F. Model evaluation in nonlinear mixed effect models, with applications to pharmacokinetics. J Soc Fr Statistique. 2010;151(1):106–28.
Holford N. The visual predictive check—superiority to standard diagnostic (Rorschach) plots. PAGE 14. 2005;Abstr 738. Available from: www.page-meeting.org/?abstract=738 .
Karlsson M, Holford N. A tutorial on Visual Predictive Checks. PAGE 17. 2008;Abstr 1434. Available from: www.page-meeting.org/?abstract=1434 .
Nguyen THT, Comets E, Mentré F. Extension of NPDE for evaluation of nonlinear mixed effect models in presence of data below the quantification limit with applications to HIV dynamic model. J Pharmacokinet Pharmacodyn. 2012;39(5):499–518.
Comets E, Brendel K, Mentré F. Computing normalised prediction distribution errors to evaluate nonlinear mixed-effect models: the npde add-on package for R. Comput Methods Prog Biomed. 2008;90(2):154–66.
MONOLIX. MOdèles NOn LInéaires à effets miXtes). Antony, France; 2016. Available from: http: //lixoft.com/products/monolix/ .
Desmée S, Mentré F, Veyrat-Follet C, Guedj J. Nonlinear mixed-effect models for prostate-specific antigen kinetics and link with survival in the context of metastatic prostate cancer: a comparison by simulation of two-stage and joint approaches. AAPS J. 2015;17(3):691–9.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29.
Roach M, Hanks G, Thames H, Schellhammer P, Shipley WU, Sokol GH, et al. Defining biochemical failure following radiotherapy with or without hormonal therapy in men with clinically localized prostate cancer: recommendations of the RTOG-ASTRO Phoenix Consensus Conference. Int J Radiat Oncol Biol Phys. 2006;65(4):965–74.
Stephenson AJ, Kattan MW, Eastham JA, Dotan ZA, Bianco FJ, Lilja H, et al. Defining biochemical recurrence of prostate cancer after radical prostatectomy: a proposal for a standardized definition. J Clin Oncol. 2006;24(24):3973–8.
Tannock IF, Fizazi K, Ivanov S, Karlsson CT, Fléchon A, Skoneczna I, et al. Aflibercept versus placebo in combination with docetaxel and prednisone for treatment of men with metastatic castration-resistant prostate cancer (VENICE): a phase 3, double-blind randomised trial. Lancet Oncol. 2013;14(8):760–8.
R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria; 2017. Available from: https://www.R-project.org/ .
Lawrence Gould A, Boye ME, Crowther MJ, Ibrahim JG, Quartey G, Micallef S, et al. Joint modeling of survival and longitudinal non-survival data: current methods and issues. Report of the DIA Bayesian joint modeling working group. Stat Med. 2015;34(14):2181–95.
Desmée S, Mentré F, Veyrat-Follet C, Sébastien B, Guedj J. Using the SAEM algorithm for mechanistic joint models characterizing the relationship between nonlinear PSA kinetics and survival in prostate cancer patients. Biometrics. 2017;73:305–12.
Cox DR, Snell EJA. general definition of residuals. J R Stat Soc Series B Stat Methodol. 1968;30(2):248–75.
Collett D. Modelling survival data in medical research. Chapman and Hall/CRC; 2014.
Andersen PK, Gill RD. Cox’s regression model for counting processes: a large sample study. Ann Stat. 1982;10(4):1100–20.
Therneau TM, Grambsch PM, Fleming TR. Martingale-based residuals for survival models. Biometrika. 1990;77(1):147–60.
Hutmacher M. A Visual Predictive Check for the evaluation of the hazard function in time-to-event analyses. PAGE 22. 2013;Abstr 2940. Available from: www.page-meeting.org/?abstract=2940.
Huh Y, Hutmacher M. Application of a hazard-based visual predictive check to evaluate parametric hazard models. J Pharmacokinet Pharmacodyn. 2016;43(1):57–71.
Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):515–26.
ACKNOWLEDGMENTS AND DISCLOSURES
Marc Cerou received funding from Institut de Recherches Internationales Servier. The authors thank Hervé Le Nagard and Francois Cohen for the use of the computer cluster services hosted on the “Centre de Biomodélisation UMR1137” and Solène Desmée for her help with setting up the simulation study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 429 kb)
Rights and permissions
About this article
Cite this article
Cerou, M., Lavielle, M., Brendel, K. et al. Development and performance of npde for the evaluation of time-to-event models. Pharm Res 35, 30 (2018). https://doi.org/10.1007/s11095-017-2291-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11095-017-2291-3