Abstract
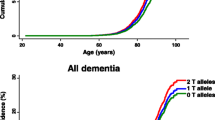
Clusterin (CLU; also known as apolipoprotein J, ApoJ) is a protein of inconstant structure known to be involved in diverse processes inside and outside of brain cells. CLU can act as a protein chaperon or protein solubilizer, lipid transporter as well as redox sensor and be anti- or proapoptotic, depending on context. Primary structure of CLU is encoded by CLU gene which contains single nucleotide polymorphisms (SNP’s) associated with the risk of late-onset Alzheimer’s disease (LOAD). Studying a sample of Czech population and using the case-control association approach we identified C allele of the SNP rs11136000 as conferring a reduced risk of LOAD, more so in females than in males. Additionally, data from two smaller subsets of the population sample suggested a possible association of rs11136000 with diabetes mellitus. In a parallel study, we found no association between rs11136000 and mild cognitive impairment (MCI). Our findings on rs11136000 and LOAD contradict those of some previous studies done elsewhere. We discuss the multiple roles of CLU in a broad range of molecular mechanisms that may contribute to the variability of genetic studies of CLU in various ethnic groups. The above discordance notwithstanding, our conclusions support the association of rs1113600 with the risk of LOAD.

Similar content being viewed by others
Abbreviations
- AD:
-
Alzheimer’s disease
- ALOX5AP:
-
Arachidonate 5-lipoxygenase-activating protein
- ApoE4:
-
Apolipoprotein E4
- ApoJ:
-
Apolipoprotein J (a.k.a. clusterin)
- BMI:
-
Body mass index
- cCLU:
-
Cytoplasmic form of clusterin
- CI:
-
Confidence interval
- CLU:
-
Clusterin
- CLU :
-
Gene encoding clusterin
- CT:
-
Computed tomography
- CVA:
-
Cerebrovascular accident
- DM:
-
Diabetes mellitus
- GWAS:
-
Genome-wide association study
- LOAD:
-
Late-onset Alzheimer’s disease
- MCI:
-
Mild cognitive impairment
- MMSE:
-
Mini-mental state examination test
- MoCA:
-
Montreal cognitive assessment
- MS:
-
Manuscript
- nCLU:
-
Nuclear form of clusterin
- OR:
-
Odds ratio
- PCR:
-
Polymerase chain reaction
- RR:
-
Risk ratio
- SBE:
-
Single base extension (reaction)
- sCLU:
-
Secretory form of clusterin
- SNP:
-
Single nucleotide polymorphisms
- TNK1:
-
Tyrosine kinase non-receptor 1
References
Gatz M, Reynolds CA, Fratiglioni L, Johanson B, Mortimer JA, Berg S, Fiske A, Pedersen L (2012) Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 32:168–174
Povová J, Ambroz P, Bar M, Pavuková V, Šerý O, Tomášková H, Janout V (2012) Epidemiological of and risk factors for Alzheimer’s disease: a review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 156:108–114. https://doi.org/10.5507/bp.2012.055
Šerý O, Povová J, Míšek I, Pešák L, Janout V (2013) Molecular mechanisms of neuropathological changes in Alzheimer’s disease: a review. Folia Neuropathol 51:1–9. https://doi.org/10.5114/fn.2013.34190
Guo T, Zhang D, Zeng Y, Huang TY, Xu H, Zhao Y (2020) Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol Neurodegener 15:40. https://doi.org/10.1186/s13024-020-00391-7
Šerý O, Povová J, Balcar VJ (2014) Perspectives in genetic prediction of Alzheimer’s disease. Neuroendocrinol Lett 35:359–366
Šerý O, Hlinecká L, Povová J, Bonczek O, Zeman T, Janout V, Ambroz P, Khan NA, Balcar VJ (2016) Arachidonate 5-lipoxygenase (ALOXAP5) gene polymorphism is associated with Alzheimer’s disease and body mass index. J Neurol Sci 362:27–32. https://doi.org/10.1016/j.jns.2016.01.022; see corrigendum in: J Neurol Sci 366:135. https://doi.org/10.1016/j.jns.2016.05.008
Šerý O, Janoutová J, Ewerlingová L, Hálová A, Lochman J, Janout V, Khan NA, Balcar VJ (2017) CD36 gene polymorphism is associated with Alzheimer disease. Biochimie 135:46–53. https://doi.org/10.1016/j.biochi.2017.01.009
Zhou L, Li H-Y, Wang J-H, Deng ZZ, Shan YL, Tan S, Shi YH, Zhang MX, Liu SX, Zhang BJ, Hong MF, Lu ZQ, Huang XM (2018) Correlation of gene polymorphisms of CD36 and ApoE with susceptibility of Alzheimer disease. A case-control study. Medicine (Baltimore) 97(38):e12470. https://doi.org/10.1097/MD.0000000000012470
Šerý O, Goswami N, Balcar VJ (2020) Chapter 4: CD36 gene polymorphisms and Alzheimer’s disease. In: Martin C, Preedy V (eds) Neuroscience of dementia: genetics, behaviour and diet in dementia. I. Genetics, molecular & cellular biology. ISBN: 9780128160435 Elsevier (© Academic Press). Published 1 Aug 2020
Hálová A, Janoutová J, Ewerlingová L, Janout V, Bonczek O, Zeman T, Gerguri T, Balcar VJ, Šerý O (2018) CHAT gene polymorphism rs3810950 is associated with the risk of Alzheimer’s disease in the Czech population. J Biomed Sci 25:41. https://doi.org/10.1186/s12929-018-0444-2
Zeman T, Balcar VJ, Cahová K, Janoutová J, Janout V, Lochman J, Šerý O (2020) Polymorphism rs11867353 of tyrosine kinase non-receptor 1 (TNK1) gene is a novel genetic marker for Alzheimer’s disease. Mol Neurobiol (in press). https://doi.org/10.1007/s12035-020-02153-4
Giri M, Zhang M, Lu Y (2016) Genes associated with Alzheimer’s disease: an overview and current status. Clin Interv Aging 11:665–681
De Roeck A, van Broekhoven C, Sleegels K (2019) The role of ABCA7 in Alzheimer’s disease: evidence from genomics, transriptomics and methylomics. Acta Neuropathol 138:201–220
Mahley RW, Rall SC (2000) Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet 1:507–537. https://doi.org/10.1146/annurev.genom.1.1.507
Stocker H, Möllers T, Perna L, Brenner H (2018) The genetic risk of Alzheimer’s disease beyond APOE ε4: systematic review of Alzheimer’s genetic risk scores. Transl Psychiatry 8:166. https://doi.org/10.1038/s41398-018-0221-8
Yao M, Fang M, Zheng W, Dong Z, Yao D (2018) Role of secretory clusterin in hepatocarcinogenesis. Transl Gastroenterol Hepatol 3:48. https://doi.org/10.21037/tgh.2018.07.13
Koltai T (2014) Clusterin: a key player in cancer chemoresistance and its inhibition. OncoTargets Ther 7:447–456. https://doi.org/10.2147/OTT.S58622
Wilson M, Zubeidi A (2017) Clusterin as a therapeutic agent. Expert Opinion Ther Targets 21:201–203
Foster EM, Dangla-Valls A, Lovestone S, Ribe EM, Buckley NJ (2019) Clusterin in Alzheimer’s disease: mechanisms, genetics, and lessons from other pathologies. Front Neurosci 13:164. https://doi.org/10.3389/fnins.2019.00164
Humphreys DT, Carver JA, Easterbrook-Smith SB, Wilson MR (1999) Clusterin has chaperone-like activity similar to that of small heat shock proteins. J Biol Chem 274:6875–6881. https://doi.org/10.1074/jbc.274.11.6875
Rohne P, Prochnow H, Wolf S, Renner B, Koch-Brandt C (2014) The chaperone activity of clusterin is dependent on glycosylation and redox environment. Cell Physiol Biochem 34:1626–1639. https://doi.org/10.1159/000366365
Rohne P, Prochnow H, Koch-Brandt C (2016) The CLU-files: disentanglement of a mystery. Biomol Concepts 7:1–15. https://doi.org/10.1515/bmc-2015-0026
Trougakos IP (2013) The molecular chaperone apolipoprotein J/clusterin as a sensor of oxidative stress: implications in therapeutic approaches—a mini-review. Gerontology 59:514–523. https://doi.org/10.1159/000351207
Park S, Mathis KW, Lee IK (2014) The physiological roles of apolipoprotein J/clusterin in metabolic and cardiovascular diseases. Rev Endocr Metab Disord 15:45–53. https://doi.org/10.1007/s11154-013-9275-3
Zhong J, Yu X, Dong X, Lu H, Zhou W, Li L, Li Z, Sun P, Shi X (2018) Therapeutic role of meloxicam targeting secretory clusterin-mediated invasion in hepatocellular carcinoma cells. Oncol Lett 15:7191–7199. https://doi.org/10.3892/ol.2018.8186
Lourda M, Trougakos IP, Gonos ES (2007) Development of resistance to chemotherapeutic drugs in human osteosarcoma cell lines largely depends on up-regulation of clusterin/apolipoprotein J. Int J Cancer 120:611–622. https://doi.org/10.1002/ijc.22327
Trougakos IP, Gonos ES (2006) Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in ageing and age-related diseases. Free Radic Res 40:1324–1334. https://doi.org/10.1080/10715760600902310
Gorini G, Harris RA, Mayfield RD (2014) Proteomic approaches and identification of novel therapeutic targets for alcoholism. Neuropsychopharmacol Rev 39:104–130. https://doi.org/10.1038/npp.2013.182
Alon U, Barkai N, Notterman DA, Gish K, Ybarr S, Mack D, Levine AJ (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci U S A 96:6745–6750
Alaylıoğlu M, Gezen-Ak D, Dursun E, Bilgiç B, Hanağası H, Ertan T, Gürvit H, Emre M, Eker E, Uysal Ö, Yılmazer S (2016) The association between clusterin and APOE polymorphisms and late-onset Alzheimer disease in a Turkish cohort. J Geriatr Psychiatry Neurol 29:221–226. https://doi.org/10.1177/0891988716640373
Jones SE, Jomary C (2002) Clusterin. Int J Biochem Cell Biol 34:427–431. https://doi.org/10.1016/s1357-2725(01)00155-8
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M et al (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41:1094–1099. https://doi.org/10.1038/ng.439
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamsher ML, Pahwa JS et al (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41:1088–1093. https://doi.org/10.1038/ng.440
Karch CM, Goate AM (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry 77:43–51. https://doi.org/10.1016/j.biopsych.2014.05.006
Kiddle SJ, Sattlecker M, Proitsi P, Simmons A, Westman E, Bazenet C et al (2014) Candidate blood proteome markers of Alzheimer’s disease onset and progression: a systematic review and replication study. J Alzheimers Dis 38:515–531. https://doi.org/10.3233/JAD-130380
McGeer PL, Kawamata T, Walker DG (1992) Distribution of clusterin in Alzheimer brain tissue. Brain Res 579:337–341
Miners JS, Clarke P, Love S (2017) Clusterin levels are increased in Alzheimer’s disease and influence the regional distribution of Aβ. Brain Pathol 27:305–313. https://doi.org/10.1111/bpa.12392
Shepherd CE, Affleck AJ, Bahar AY, Crew-Jones F, Halliday GM (2019) Intracellular and secreted forms of clusterin are elevated early in Alzheimer’s disease and associate with both Aβ and tau pathology. Neurobiol Aging 89:129–131. https://doi.org/10.1016/j.neurobiolaging.2019.10.025
Calero M, Rostagno A, Matsubara E, Zlokovic B, Frangione B, Ghiso J (2000) Apolipoprotein J (clusterin) and Alzheimer’s disease. Microsc Res Tech 50:305–315. https://doi.org/10.1002/1097-0029(20000815)50:4<305::AID-JEMT10>3.0.CO;2-L
Zhou Y, Hayashi I, Wong J, Tugusheva K, Renger JJ, Zerbinatti C (2014) Intracellular clusterin interacts with brain isoforms of the bridging integrator 1 and with the microtubule-associated protein tau in Alzheimer’s disease. PLoS One 9(7):e103187
Kashem MA, Sultana N, Balcar VJ (2018) Exposure of rat neural stem cells to ethanol affects cell numbers and alters expression of 28 proteins. Neurochem Res 43:1841–1854. https://doi.org/10.1007/s11064-018-2600-1
Kashem MA, Ahmed S, Sultana N, Ahmed EU, Pickford R, Rae C, Šerý O, McGregor IS, Balcar VJ (2016) Metabolomics of neurotransmitters and related metabolites in post-mortem tissue from the dorsal and ventral striatum of alcoholic postmortem human brain. Neurochem Res 41:385–397. https://doi.org/10.1007/s11064-015-1605-2
Kashem MA, Šerý O, Pow DL, Rowlands B, Rae CD, Balcar VJ (2020) Actions of alcohol in brain: genetics, metabolomics, GABA receptors, proteomics and glutamate transporter GLAST/EAAT1. Curr Mol Pharmacol 13 (in press). https://doi.org/10.2174/1874467213666200424155244
Kamboh MI, Minster RL, Demirci FY, Ganguli M, DeKosky ST, Lopez OL, Barmada MM (2012) Association of CLU and PICALM variants with Alzheimer’s disease. Neurobiol Aging 33:518–521. https://doi.org/10.1016/j.neurobiolaging.2010.04.015
Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F et al (2010) Replication of CLU, CR1, and PICALM associations with Alzheimer’s disease. Arch Neurol 67:961–964. https://doi.org/10.1001/archneurol.2010.147
Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K et al (2006) Mild cognitive impairment. Lancet 367(9518):1262–1270. https://doi.org/10.1016/S0140-6736(06)68542-5
R Development Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Seripa D, Panza F, Paroni G, D’Onofrio G, Bisceglia P, Gravina C et al (2018) Role of CLU, PICALM and TNK1 genotypes in aging with and without Alzheimer’s disease. Mol Neurobiol 55:4333–4344. https://doi.org/10.1007/s12035-017-0547-x
Zhu R, Liu X, He Z (2018) Association between CLU gene rs11136000 polymorphism and Alzheimer’s disease: an updated meta-analysis. Neurol Sci 39:679–689. https://doi.org/10.1007/s10072-018-3259-8
Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M et al (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. J Amer Med Assoc 303:1832–1840. https://doi.org/10.1001/jama.2010.574
Ling I-F, Bhongsatiern J, Simpson JF, Fardo DW, Estus S (2012) Genetics of clusterin isoform expression and Alzheimer’s disease risk. PLoS One 7(4):e33923
Golenkina SA, Gol’tsov AI, Kuznetsov IL, Grigorenko AP, Andreeva TV, Reshetov DA et al (2010) Analysis of clusterin gene (CLU/APOJ) polymorphism in Alzheimer’s disease patients and in normal cohorts from Russian populations (in Russian). Mol Biol (Moscow) 44:620–626
Carrasquillo MM, Crook JE, Pedraza O, Thomas CS, Pankratz VS, Allen M et al (2015) Late-onset Alzheimer’s risk variants in memory decline, incident mild cognitive impairment and Alzheimer’s disease. Neurobiol Aging 36:60–67. https://doi.org/10.1016/j.neurobiolaging.2014.07.042
Sweet RA, Seltman H, Emanuel JE, Lopez OL, Becker JT, Bis JC et al (2012) Effect of Alzheimer’s disease risk genes on trajectories of cognitive function in the cardiovascular health study. Am J Psychiatry 169:954–962. https://doi.org/10.1176/appi.ajp.2012.11121815
Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG et al (2013) Alzheimer risk variant CLU and brain function during aging. Biol 73:399–405. https://doi.org/10.1016/j.biopsych.2012.05.026
Herring SK, Moon HJ, Rawal P, Chhibber A, Zhao L (2019) Brain clusterin protein isoforms and mitochondrial localization. Elife 8:e48255. https://doi.org/10.7554/eLife.48255
Kim N, Choi WS (2011) Proapoptotic role of nuclear clusterin in brain. Anat Cell Biol 44:169–175. https://doi.org/10.5115/acb.2011.44.3.169
Wyatt AR, Yerbury JJ, Berghofer P, Greguric I, Katsifis A, Dobson CM, Wilson MR (2011) Clusterin facilitates in vivo clearance of extracellular misfolded proteins. Cell Mol Life Sci 68:3919–3931
Aghajanpour-Mir M, Amjadi-Moheb F, Dadkhah T, Hosseini SR, Ghadami E, Assadolahi E, Akhavan-Niaki H, Ahangar AA (2019) Informative combination of CLU rs11136000, serum HDL levels, diabetes and age as a new piece of puzzle-picture of predictive medicine for cognitive disorders. Mol Biol Rep 46:1033–1041. https://doi.org/10.1007/s11033-018-4561-5
Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. https://doi.org/10.1093/nar/gky1141
Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217. https://doi.org/10.1016/j.cell.2006.07.031
Gregory JM, Whiten DR, Brown RA, Barros TP, Kumita JR, Yerbury JJ et al (2017) Clusterin protects neurons against intracellular proteotoxicity. Acta Neuropathol Commun 5:81
Xu Z, Lee A, Nouwens A, Henderson RD, McCombe PA (2018) Mass spectroscopy analysis of plasma from amyotrophic lateral sclerosis and control subjects. Amyotroph Lat Scler Frontotemp Degener 19:362–376. https://doi.org/10.1080/21678421.2018.1433689
Mohanty L, Henderson RD, McCombe PA, Lee A (2020) Levels of clusterin, CD5L, ficolin-3 and gelsolin in ALS patients and controls. Amyotroph Lat Scler Frontotemp Degener. https://doi.org/10.1080/21678421.2020.1779303
Matukumalli SR, Tangirala R, Rao CM (2017) Clusterin: full-length protein and one of its chains show opposing effects on cellular lipid accumulation. Sci Rep 7:41235. https://doi.org/10.1038/srep41235
Scheidt T, Łapińska U, Kumita JR, Whiten DR, Klenerman D, Wilson MR et al (2019) Secondary nucleation and elongation occur at different sites on Alzheimer’s amyloid-β aggregates. Sci Adv 5(4):eaau3112. https://doi.org/10.1126/sciadv.aau3112
Picón-Pagès P, Bonet J, García-García J, Garcia-Buendia J, Gutierrez D, Valle J (2019) Human albumin impairs amyloid β-peptide fibrillation through its C-terminus: from docking modeling to protection against neurotoxicity in Alzheimer’s disease. Comput Struct Biotechnol J 17:963–971. https://doi.org/10.1016/j.csbj.2019.06.017
Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR (2007) The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillary structures. FASEB J 21:2312–2322
Martin-Rehrmann MD, Hoe HS, Capuani EM, Rebeck GW (2005) Association of apolipoprotein J-positive beta-amyloid plaques with dystrophic neurites in Alzheimer’s disease brain. Neurotox Res 7:231–242. https://doi.org/10.1007/BF03036452
Satapathy S, Dabba RA, Wilson MR (2020) Rapid high-yield expression and purification of fully post-translationally modified recombinant clusterin and mutants. Sci Rep 10:14243. https://doi.org/10.1038/s41598-020-70990-3
Qiu L, He Y, Tang H, Zhou Y, Wang J, Zhang W et al (2016) Genetically-mediated grey and white matter alteration in normal elderly individuals with the CLU-C allele gene. Curr Alzheimer Res 13:1302–1310. https://doi.org/10.2174/1567205013666160703180531
Šerý O, Hlinecká L, Balcar VJ, Janout V, Povová J (2014) Diabetes, hypertension and stroke—does Alzheimer protect you? Neuroendocrinol Lett 35:691–696
Langenberg C, Hardy R, Kuh D, Wadsworth EJ (2003) Influence of height, leg and trunk length on pulse pressure, systolic and diastolic blood pressure. J Hypertens 21:537–543. https://doi.org/10.1097/01.hjh.0000052472.40108.4e
Stone J, Jonstone DM, Mitrofanis J, O’Rourke M (2015) The mechanical cause of age-related dementia (Alzeimer’s disease): the brain is destroyed by the pulse. J Alzheimer’s Dis 44:355–373. https://doi.org/10.3233/JAD-141884
Levin RA, Carnegie MH, Celermajer DS (2020) Pulse pressure: an emerging therapeutic target for dementia. Front Neurosci 14:a669. https://doi.org/10.3389/fnins.2020.00669
Trougakos IP, Poulakou M, Stathotos M, Chalikia A, Melidonis A, Gonos ES (2002) Serum levels of the senescence biomarker clusterin/apolipoprotein J increase significantly in diabetes type II and during development of coronary heart disease or at myocardial infarction. Exp Gerontol 37:1175–1187
Seo JA, Kang MC, Ciaraldi TP, Kim SS, Park KS, Choe C et al (2018) Circulating ApoJ is closely associated with insulin resistance in human subjects. Metabolism 78:155–166
Gelissen IC, Hochgrebe T, Wilson MR, Easterbrook-Smith SB, Jessup W, Dean RT et al (1998) Apolipoprotein J (clusterin) induces cholesterol export from macrophage-foam cells: a potential anti-atherogenic function? Biochem J 331:231–237. https://doi.org/10.1016/S0021-9150(97)89866-8
Zandl-Lang M, Fanaee-Danesh E, Sun Y, Albrecher N, Gali CC (2018) Regulatory effects of simvastatin and ApoJ on APP processing and amyloid-β clearance in blood-brain barrier endothelial cells. Biochim Biophys Acta Mol Cell Biol Lipids 1863:40–60. https://doi.org/10.1016/j.bbalip.2017.09.008
Cai R, Han J, Sun J, Huang R, Tian S, Shen Y, Dong X, Xia W, Wang S (2016) Plama clusterin and the CLU gene rs11136000 variant are associated with mild cognitive impairment in type 2 diabetic patients. Front Aging Neurosci 8:a179. https://doi.org/10.3389/fnagi.2016.00179
Acknowledgements
We would like to thank to Petr Ambroz and Ondřej Machaczka for organizing the collection of samples.
Funding
This work has been supported by Czech Health Research Council, Czech Republic (AZV CR)—grant projects No. NV18-04-00455 and 16-29900A.
Author information
Authors and Affiliations
Contributions
VJB—writing, interpretation of results, final MS preparation; TZ—statistical analyses of data and interpretation, writing; JL—selection and design of laboratory methods, genetic analysis; VJ—conceptualisation, funding, patient enrolment; JJ—collection and curation of data; OS—conceptualisation, funding, laboratory analysis.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflicts of interest.
Consent to Publish
All authors have read and approved the final version of the manuscript.
Ethical Approval
Informed consent was obtained from all participants or their legal representatives and formal approval for the study was granted by the Ethics Committee of the Faculty of Medicine, University of Ostrava, Czech Republic and by the Ethics Committee of St. Anne’s University Hospital, Brno, Czech Republic. The projects “Genetics and Epidemiology of Alzheimer’s Disease” (EK no. 11/2015), “Genetics and Epidemiology of Mild Cognitive Disorder” (EK no. 12/2015) were approved by the Committee on 16th June 2015 and project “The role of CD36 gene in pathogenesis of Alzheimer’s disease “was approved by the Ethics Committee of St. Anne’s University Hospital on 29th June 2017.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Balcar, V.J., Zeman, T., Janout, V. et al. Single Nucleotide Polymorphism rs11136000 of CLU Gene (Clusterin, ApoJ) and the Risk of Late-Onset Alzheimer’s Disease in a Central European Population. Neurochem Res 46, 411–422 (2021). https://doi.org/10.1007/s11064-020-03176-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-020-03176-y