Abstract
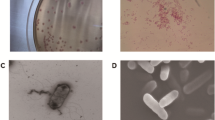
Candia (Starmera) stellimalicola is a yeast species spread worldwide and recovered from varieties of ecological reservoirs, but human infections are rarely reported. In this study, we reported an intra-abdominal infection case caused by C. stellimalicola and described its microbiological and molecular characteristics. C. stellimalicola strains were isolated from ascites fluid of an 82-year-old male patient having diffuse peritonitis with fever and elevated WBC counts. Routine biochemical and MALDI-TOF MS methods failed to identify the pathogenic strains. Phylogenetic analysis of 18S, 26S and internal transcribed space (ITS) rDNA regions, as well as whole-genome sequence identified the strains as C. stellimalicola. Compared with other Starmera species, C. stellimalicola had unique physiological characteristics including thermal tolerance (able to grow at 42 °C), which may prompt its environmental adaptability and potential for opportunistic human infection. Fluconazole minimum inhibitory concentration (MIC) values of the strains identified in this case was 2 mg/L, and the patient had a favorable outcome after receiving fluconazole treatment. In comparison, the majority of C. stellimalicola strains previously documented had high MIC values (≥ 16 mg/L) to fluconazole. In conclusion, with the raise in human infections caused by rare fungal pathogens, molecular diagnostic remains the most efficient way for accurate species identification; and antifungal susceptibility testing is essential to guide proper patient management.

Similar content being viewed by others
Data Availability
Sequences of the rDNA regions of C. stellimalicola strain HLJSST01 obtained in this study have been deposited in NCBI GenBank database under accession nos. OQ745670 (for 18S rDNA), OQ745669 (for 26S rDNA) and OQ750115 (for ITS region). In addition, the strains’ next-generation sequencing data for raw reads and genome assembly is available at NMDC database under Bioproject accession no. PRJNA952807.
References
World Health Organization. WHO fungal priority pathogens list to guide research, development and public health action. Geneva 2022.
Fisher MC, et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio. 2020;11(3):e00449-e520. https://doi.org/10.1128/mBio.00449-20.
Pappas PG, et al. Invasive candidiasis. Nat Rev Dis Primers. 2018;4:18026. https://doi.org/10.1038/nrdp.2018.26.
Yamada Y, et al. The phylogeny of strains of species of the genus Pichia Hansen (Saccharomycetaceae) based on the partial sequences of 18S ribosomal RNA: the proposals of Phaffomyces and Starmera, the new genera. Bull Fac Agric Shizuoka Univ (Japan). 1998;47:27–35.
Moreira JD, et al. Starmera foglemanii sp. nov. and Starmera ilhagrandensis sp. nov., two novel yeast species isolated from ephemeral plant substrates. Int J Syst Evol Microbiol. 2020;70(7):4378–83. https://doi.org/10.1099/ijsem.0.004300.
Suzuki M, Nakase T, Komagata K. Candida stellimalicola, a new species of anamorphic yeast isolated from star apple in Thailand. J Gen Appl Microbiol. 1994;40(2):115–21. https://doi.org/10.2323/jgam.40.115.
Nguyen NH, Suh SO, Blackwell M. Five novel Candida species in insect-associated yeast clades isolated from Neuroptera and other insects. Mycologia. 2007;99(6):842–58. https://doi.org/10.3852/mycologia.99.6.842.
Brandao LR, et al. Diversity and antifungal susceptibility of yeasts isolated by multiple-tube fermentation from three freshwater lakes in Brazil. J Water Health. 2010;8(2):279–89. https://doi.org/10.2166/wh.2009.170.
Dupont D, et al. Donor derived Candida stellimalicola in a clinical specimen: preservation fluid contamination during pancreas procurement. Mycopathologia. 2018;183(3):573–7. https://doi.org/10.1007/s11046-017-0171-3.
Borman AM, et al. Fluconazole resistance in isolates of uncommon pathogenic yeast species from the United Kingdom. Antimicrob Agents Chemother. 2019;63(8):e00211-e219. https://doi.org/10.1128/AAC.00211-19.
Raja HA, et al. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod. 2017;80(3):756–70. https://doi.org/10.1021/acs.jnatprod.6b01085.
Kumar S, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096.
Fan X, et al. A pseudo-outbreak of Cyberlindnera fabianii funguria: Implication from whole genome sequencing assay. Front Cell Infect Microbiol. 2023;13:1130645. https://doi.org/10.3389/fcimb.2023.1130645.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. https://doi.org/10.1093/bioinformatics/btp324.
Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. https://doi.org/10.1093/bioinformatics/btp352.
McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. https://doi.org/10.1101/gr.107524.110.
Jain C, et al. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114. https://doi.org/10.1038/s41467-018-07641-9.
Stanke M, et al. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24(5):637–44. https://doi.org/10.1093/bioinformatics/btn013.
Daniel HM, et al. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 2009;9(5):774–83. https://doi.org/10.1111/j.1567-1364.2009.00520.x.
Jatmiko YD, Lopes MDB, Barton MD. Molecular identification of yeasts isolated from Dadih by RFLP-PCR and assessment on their ability in utilizing lactate. Microbiol Indones. 2012;6(1):5. https://doi.org/10.5454/mi.6.1.5.
Freitas LFD, et al. Starmera pilosocereana sp. nov., a yeast isolated from necrotic tissue of cacti in a sandy coastal dune ecosystem. Int J Syst Evol Microbiol. 2015;65(12):4474–8.
Kurtzman CP, Robnett CJ, Basehoar-Powers E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 2008;8(6):939–54. https://doi.org/10.1111/j.1567-1364.2008.00419.x.
Garcia-Bustos V, et al. Climate change, animals, and Candida auris: insights into the ecological niche of a new species from a one health approach. Clin Microbiol Infect. 2023. https://doi.org/10.1016/j.cmi.2023.03.016.
Casadevall A, Kontoyiannis DP, Robert V. On the emergence of Candida auris: climate change, azoles, swamps, and birds. MBio. 2019. https://doi.org/10.1128/mBio.01397-19.
Funding
This work was supported by the National Key Research and Development Program of China (Grant nos. 2022YFC2303002 and 2022YFC2504800).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant conflict of interest.
Additional information
Handling Editor: Nancy Wengenack.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, X., Dai, RC., Yan, XF. et al. Clinical, Microbiological, and Molecular Characterization of Candia (Starmera) stellimalicola, a Rare Fungal Pathogen Causing Human Infections. Mycopathologia 188, 345–352 (2023). https://doi.org/10.1007/s11046-023-00752-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-023-00752-y