Abstract
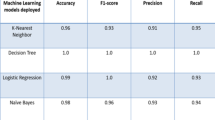
In this study, we analyzed the performance of the morphological features (MF) and morphological distance-related features (MDRF) in the classification of autism spectrum disorder (ASD) and typical development (TD). Initially, we preprocessed the structural magnetic resonance images (sMRI) of ASD and TD from seven sites publicly available in the autism brain imaging data exchange (ABIDE-I and ABIDE-II) database using a standard pipeline. Further, sMRI images were parcellated into different regions using the Destrieux atlas. Moreover, MF (surface area) and MDRF were calculated from each region. We tested the performance of the MF and MDRF on each site by feeding them to classifiers such as random forest (RF), support vector machines (SVM), and multi-layer perceptron (MLP). Our results suggested that the MDRF could classify the ASD and TD better than the MF. Furthermore, RF gave a single-site average classification accuracy of 91.78% and 95.27% using MF and MDRF, respectively. We achieved the average classification accuracy of 69.08% and 82.91% between the sites using MF and MDRF, respectively. Our results suggested that the frontal lobe and right hemisphere contributed more MDRF to the machine learning model. Furthermore, in the USM site, many features were found to be from the frontal lobe (15 distance features) and frontal-parietal (11 distance features) lobe. The results suggest that the MDRF can be a valuable feature metric for classifying ASD-like neurodevelopmental disorders.









Similar content being viewed by others
Data availability
The data used for this study was obtained from the publicly available ABIDE database https://fcon_1000.projects.nitrc.org/indi/abide/.
References
American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders, 5th ed. American Psychiatric Publishing, 2013. https://doi.org/10.1176/-appi.books.9780890425787
Karimi P, Kamali E, Mousavi SM, Karahmadi M (2017) Environmental factors influencing the risk of autism. J Res Med Sci 22:27. https://doi.org/10.4103/1735-1995.200272
Zeidan J, Fombonne E, Scorah J, Ibrahim A, Durkin MS, Saxena S, Yusuf A, Shih A, Elsabbagh M (2022) Global prevalence of autism: A systematic review update. Autism Res: Official J Int Soc Autism Res 15(5):778–790. https://doi.org/10.1002/aur.2696
Subramanyam AA, Mukherjee A, Dave M, Chavda K (2019) Clinical practice guidelines for autism spectrum disorders. Indian J Psychiatry 61(Suppl 2):254–269. https://doi.org/10.4103/psychiatry.IndianJPsychiatry_542_18
Murphy CM, Wilson CE, Robertson DM, Ecker C, Daly EM, Hammond N, Galanopoulos A, Dud I, Murphy DG, McAlonan GM (2016) Autism spectrum disorder in adults: diagnosis, management, and health services development. Neuropsychiatr Dis Treat 12:1669–1686. https://doi.org/10.2147/-NDT.S65455
Pagnozzi AM, Conti E, Calderoni S, Fripp J, Rose SE (2018) A systematic review of structural MRI biomarkers in autism spectrum disorder: A machine learning perspective. Int J Develop Neurosci: Official J Int Soc Develop Neurosci 71:68–82. https://doi.org/10.1016/j.ijdevneu.2018.08.010
Torun YT, Güney E, Iseri E (2015) Structural and functional brain imaging in autism spectrum disorders. INTECH. https://doi.org/10.5772/59121
Ismail MM, Keynton RS, Mostapha MM, ElTanboly AH, Casanova MF, Gimel’farb GL, El-Baz A (2016) Studying autism spectrum disorder with structural and diffusion magnetic resonance imaging: A Survey. Front Hum Neurosci 10:211. https://doi.org/10.3389/fnhum.2016.00211
O'Neill M, Shear T (2018) EEG for diagnosis of autism spectrum disorder. Pediatr Neurol Briefs. 32. 13. https://doi.org/10.15844/pedneurbriefs-32-13
Katuwal GJ, Cahill ND, Baum SA, Michael AM (2015). The predictive power of structural MRI in Autism diagnosis. Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual International Conference, 2015, 4270–4273. https://doi.org/10.1109/EMBC.2015.7319338
Kucharsky Hiess R, Alter R, Sojoudi S, Ardekani BA, Kuzniecky R, Pardoe HR (2015) Corpus callosum area and brain volume in autism spectrum disorder: quantitative analysis of structural MRI from the ABIDE database. J Autism Dev Disord 45(10):3107–3114. https://doi.org/10.1007/s10803-015-2468-8
McAlonan GM, Cheung V, Cheung C, Suckling J, Lam GY, Tai KS, Yip L, Murphy DG, Chua SE (2005). Mapping the brain in autism. A voxel-based MRI study of volumetric differences and intercorrelations in autism. Brain : a journal of neurology, 128(Pt 2):268–276. https://doi.org/10.1093/brain/awh332
Rojas DC, Peterson E, Winterrowd E, Reite ML, Rogers SJ, Tregellas JR (2006) Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry 6:56. https://doi.org/10.1186/1471-244X-6-56
McAlonan GM, Daly E, Kumari V, Critchley HD, van Amelsvoort T, Suckling J, Simmons A, Sigmundsson T, Greenwood K, Russell A, Schmitz N, Happe F, Howlin P, Murphy DG (2002) Brain anatomy and sensorimotor gating in Asperger’s syndrome. Brain : J Neurol 125(Pt 7):1594–1606. https://doi.org/10.1093/brain/awf150
Waiter GD, Williams JH, Murray AD, Gilchrist A, Perrett DI, Whiten A (2004) A voxel-based investigation of brain structure in male adolescents with autistic spectrum disorder. Neuroimage 22(2):619–625. https://doi.org/10.1016/j.neuroimage-.2004.02.029
Akshoomoff N, Lord C, Lincoln AJ, Courchesne RY, Carper RA, Townsend J, Courchesne E (2004) Outcome classification of preschool children with autism spectrum disorders using MRI brain measures. J Am Acad Child Adolesc Psychiatry 43(3):349–357. https://doi.org/10.1097/00004583-200403000-00018
Herbert MR, Ziegler DA, Makris N, Filipek PA, Kemper TL, Normandin JJ, Sanders HA, Kennedy DN, Caviness VS Jr (2004) Localization of white matter volume increase in autism and developmental language disorder. Ann Neurol 55(4):530–540. https://doi.org/10.1002/ana.20032
Ashburner J, Friston KJ (2000) Voxel-based morphometry–the methods. Neuroimage 11(6 Pt 1):805–821. https://doi.org/10.1006/nimg.2000.0582
Jiao Y, Lu Z (2011) Predictive models for autism spectrum disorder based on multiple cortical features," 2011 Eighth International Conference on Fuzzy Systems and Knowledge Discovery (FSKD), Shanghai, China, 1611–1615. https://doi.org/10.1109/FSKD.2011.6019825
Libero LE, Schaer M, Li DD, Amaral DG, Nordahl CW (2019) A longitudinal study of local gyrification index in young boys with autism spectrum disorder. Cerebral cortex (New York, N.Y. : 1991) 29(6):2575–2587. https://doi.org/10.1093/cercor/bhy126
Boedhoe PSW, Van Rooij D, Hoogman M, Twisk JWR, Schmaal L, Abe Y, Alonso P, Ameis SH, Anikin A, Anticevic A et al (2020) Subcortical brain volume, regional cortical thickness, and cortical surface area across disorders: findings from the ENIGMA ADHD, ASD, and OCD working groups. Am J Psychiatry 177(9):834–843. https://doi.org/10.1176/appi.ajp.2020.19030331
Wang XH, Jiao Y, Li L (2018) Mapping individual voxel-wise morphological connectivity using wavelet transform of voxel-based morphology. PLoS ONE 13(7):e0201243. https://doi.org/10.1371/journal.pone.0201243
Mensen VT, Wierenga LM, van Dijk S, Rijks Y, Oranje B, Mandl RC, Durston S (2016) Development of cortical thickness and surface area in autism spectrum disorder. NeuroImage Clinical 13:215–222. https://doi.org/10.1016/-j.nicl.2016.12.003
Rahman MM, Usman OL, Muniyandi RC, Sahran S, Mohamed S, Razak RA (2020) A review of machine learning methods of feature selection and classification for autism spectrum disorder. Brain Sci 10(12):949. https://doi.org/10.3390/brainsci10120949
Thabtah F, Abdelhamid N, Peebles D (2019) A machine learning autism classification based on logistic regression analysis. Health Inform Sci Systems 7(1):12. https://doi.org/10.1007/s13755-019-0073-5
Ecker C, Rocha-Rego V, Johnston P, Mourao-Miranda J, Marquand A, Daly EM, Brammer MJ, Murphy C, Murphy DG, MRC AIMS Consortium (2010) Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. NeuroImage 49(1):44–56. https://doi.org/10.1016/j.neuroimage.2009.08.024
Xiao X, Fang H, Wu J, Xiao C, Xiao T, Qian L, Liang F, Xiao Z, Chu KK, Ke X (2017) Diagnostic model generated by MRI-derived brain features in toddlers with autism spectrum disorder. Autism Res: Official J Int Soc Autism Res 10(4):620–630. https://doi.org/10.1002/aur.1711
Alsaade FW, Alzahrani MS (2022) Classification and detection of autism spectrum disorder based on deep learning algorithms. Comput Intell Neurosci 2022:8709145. https://doi.org/10.1155/2022/8709145
Swain D, Mehta U, Bhatt A, Patel H, Patel K, Mehta D, Acharya B et al (2023) A robust chronic kidney disease classifier using machine learning. Electronics 12(1):212. https://doi.org/10.3390/electronics12010212
Umamaheswari K, Parthiban L (2021) Hybridization of cooperative particle swarm optimization with multilayer perceptron for autistic spectrum disorder diagnosis and classification. J Comput Theor Nanosci 18(3):722–731. https://doi.org/10.1166/jctn.2021.9671
Di Martino A, Yan C-G, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M (2014) The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 19(6):659–667. https://doi.org/10.1038/mp.2013.78
Di Martino A, O’connor D, Chen B, Alaerts K, Anderson JS, Assaf M, Balsters JH, Baxter L, Beggiato A, Bernaerts S (2017) Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci Data 4(1):1–15. https://doi.org/10.1038/sdata.2017.10
Fischl B (2012) FreeSurfer Neuroimage 62(2):774–781. https://doi.org/10.1016/-j.neuroimage.2012.01.021
Destrieux C, Fischl B, Dale A, Halgren E (2010) Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 53(1):1–15. https://doi.org/10.1016/j.neuroimage.2010.06.010
Chen T, Guestrin C (2016). XGBoost: A scalable tree boosting system. 785–794. https://doi.org/10.1145/2939672.2939785
Alsahaf A, Petkov N, Shenoy V, Azzopardi G (2022) A framework for feature selection through boosting. Expert Syst Appl 187:115895–2022. https://doi.org/10.1016/j.eswa.2021.115895
Luckner M, Topolski B, Mazurek M (2017). Application of XGBoost algorithm in fingerprinting localisation task. In: Saeed K, Homenda W, Chaki R (eds) Computer Information Systems and Industrial Management. CISIM 2017. Lecture Notes in Computer Science(). 10244. Springer, Cham. https://doi.org/10.1007/978-3-319-59105-6_57
Gardner MW, Dorling SR (1998) Artificial neural networks (the multilayer perceptron) - a review of applications in the atmospheric sciences. Atmos Environ 32(14–15):2627–2636. https://doi.org/10.1016/S1352-2310(97)00447-0
Boser BE, Guyon IM, Vapnik VN (1992) A training algorithm for optimal margin classifiers. In: Proceedings of the fifth annual workshop on computational learning theory. pp 144–152. https://doi.org/10.1145/130385.130401
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Pal M (2005) Random forest classifier for remote sensing classification. Int J Remote Sens 26(1):217–222. https://doi.org/10.1080/0143116041233-1269698
Kong Y, Gao J, Xu Y, Pan Y, Wang J, Liu J (2019) Classification of autism spectrum disorder by combining brain connectivity and deep neural network classifier. Neurocomputing 324:63–68. https://doi.org/10.1016/j.neucom.-2018.04.080
Wang XH, Jiao Y, Li L (2018) Diagnostic model for attention-deficit hyperactivity disorder based on interregional morphological connectivity. Neurosci Lett 685:30–34. https://doi.org/10.1016/j.neulet.2018.07.029
Khundrakpam BS, Lewis JD, Jeon S, Kostopoulos P, Itturia Medina Y, Chouinard-Decorte F, Evans AC (2019). Exploring Individual Brain Variability during Development based on Patterns of Maturational Coupling of Cortical Thickness: A Longitudinal MRI Study. Cerebral cortex (New York, N.Y. : 1991), 29(1):178–188. https://doi.org/10.1093/cercor/bhx317
Gupta V, Manoj G, Bhattacharya A, Singh Sengar S, Mishra R, Kar BR, Srivastava C, Agastinose Ronickom JF (2023) A Framework to Diagnose Autism Spectrum Disorder Using Morphological Connectivity of sMRI and XGBoost. Stud Health Technol Inform 309:33–37. https://doi.org/10.3233/SHTI230734
Mishra M, Pati UC (2021) Autism spectrum disorder detection using surface morphometric feature of sMRI in machine learning, 2021 8th International Conference on Smart Computing and Communications (ICSCC), Kochi, Kerala, India, 17–20. https://doi.org/10.1109/ICSCC51209.2021.9528240
Ali MT, ElNakieb Y, Elnakib A, Shalaby A, Mahmoud A, Ghazal M, Yousaf J, Abu Khalifeh H, Casanova M, Barnes G, El-Baz A (2022) The Role of Structure MRI in Diagnosing Autism. Diagnostics (Basel, Switzerland) 12(1):165. https://doi.org/10.3390/diagnostics12010165
Ali MT, ElNakieb Y, Elnakib A, Shalaby A, Mahmoud A, Switala A, Ghazal M, Khelifi A, Fraiwan L, Barnes G, El-Baz A (2021). Autism classification using SMRI: A recursive features selection based on sampling from multi-level high dimensional spaces. 267–270. https://doi.org/10.1109/ISBI48211.2021.9433973
Duan Y, Zhao W, Luo C, Liu X, Jiang H, Tang Y, Liu C, Yao D (2022) Identifying and predicting autism spectrum disorder based on multi-site structural MRI with machine learning. Front Hum Neurosci 15:765517. https://doi.org/10.3389/fnhum.2021.765517
Seidlitz J, Váša F, Shinn M, Romero-Garcia R, Whitaker KJ, Vértes PE, Wagstyl K, Kirkpatrick Reardon P, Clasen L, Liu S et al (2018) Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron 97(1):231-247.e7. https://doi.org/10.1016/j.neuron.2017.11.039
Zou R, Li L, Zhang L, Huang G, Liang Z, Zhang Z (2021) Predicting individual pain thresholds from morphological connectivity using structural MRI: A multivariate analysis study. Front Neurosci 15:615944. https://doi.org/10.3389/fnins.2021.615944
Li J, Kong XZ (2017) Morphological connectivity correlates with trait impulsivity in healthy adults. PeerJ 5:e3533. https://doi.org/10.7717/peerj.3533
Wang H, Jin X, Zhang Y, Wang J (2016) Single-subject morphological brain networks: connectivity mapping, topological characterization and test-retest reliability. Brain and behavior 6(4):e00448. https://doi.org/10.1002/brb3.448
Lee SH, Maenner MJ, Heilig CM (2019) A comparison of machine learning algorithms for the surveillance of autism spectrum disorder. PLoS ONE 14(9):e0222907. https://doi.org/10.1371/journal.pone.0222907
Crucitti J, Hyde C, Enticott PG, Stokes MA (2022) A systematic review of frontal lobe volume in autism spectrum disorder revealing distinct trajectories. J Integrative Neurosci 21(2):5. https://doi.org/10.31083/j.jin2102057
Carper RA, Moses P, Tigue ZD, Courchesne E (2002) Cerebral lobes in autism: early hyperplasia and abnormal age effects. Neuroimage 16(4):1038–1051. https://doi.org/10.1006/nimg.2002.1099
Bhattacharya A, Manoj G, Gupta V, Shaik Gadda AA, Vedantham D, Prince A, Rani P, Ramaniharan A, Fredo J (n.d.) Comparative evaluation of geometrical, Zernike moments, and volumetric features of the corpus callosum for discrimination of ASD using machine learning algorithms. (in press) International Journal of Biomedical Engineering and Technology. https://doi.org/10.1504/-IJBET.2022.10054054
Jain V, Selvaraj A, Mittal R, Rani P, Kilpattu Ramaniharan A, Agastinose Ronickom JF (2022) Automated diagnosis of autism spectrum disorder condition using shape based features extracted from brainstem. Stud health Technol Inform 294:53–57. https://doi.org/10.3233/SHTI220395
Fu Y, Zhang J, Li Y, Shi J, Zou Y, Guo H, Li Y, Yao Z, Wang Y, Hu B (2021) A novel pipeline leveraging surface-based features of small subcortical structures to classify individuals with autism spectrum disorder. Prog Neuropsychopharmacol Biol Psychiatry 104:109989. https://doi.org/10.1016/j.pnpbp.2020.109989
Raamana PR, Strother SC, Biomarkers AI, Lifestyle Flagship Study of Ageing, for the Alzheimer’s Disease Neuroimaging Initiative (2020) Does size matter? The relationship between predictive power of single-subject morphometric networks to spatial scale and edge weight. Brain Struct Funct 225(8):2475–2493. https://doi.org/10.1007/s00429-020-02136-0
Chen T, Chen Y, Yuan M, Gerstein M, Li T, Liang H, Froehlich T, Lu L (2020) The development of a practical artificial intelligence tool for diagnosing and evaluating autism spectrum disorder: Multicenter study. JMIR Med Inform 8(5):e15767. https://doi.org/10.2196/15767
Mishra M, Pati UC (2023) A classification framework for autism spectrum disorder detection using sMRI: Optimizer based ensemble of deep convolution neural network with on-the-fly data augmentation. Biomed Signal Process Control 104686, 2023. https://doi.org/10.1016/j.bspc.2023.104686
Gao K, Fan Z, Su J, Zeng LL, Shen H, Zhu J, Hu D (2022). Deep transfer learning for cerebral cortex using area-preserving geometry mapping. Cerebral cortex (New York, N.Y. : 1991. 32(14), 2972–2984. https://doi.org/10.1093/cercor/-bhab394
Devika K, Mahapatra D, Subramanian R, Murthy OVR (2022) Outlier-based autism detection using longitudinal structural MRI. IEEE Access 10:27794–27808. https://doi.org/10.1109/ACCESS.2022.3157613
Wang Z, Peng D, Shang Y, Gao J (2021) Autistic spectrum disorder detection and structural biomarker identification using self-attention model and individual-level morphological covariance brain networks. Front Neurosci 15:756868. https://doi.org/10.3389/fnins.2021.756868
Gao J, Chen M, Li Y, Gao Y, Li Y, Cai S, Wang J (2021) Multisite autism spectrum disorder classification using convolutional neural network classifier and individual morphological brain networks. Front Neurosci 14:629630. https://doi.org/10.3389/fnins.2020.629630
Acknowledgements
This work was supported by the Param Shivay supercomputing center of the Indian Institute of Technology (BHU), Varanasi.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: Amalin Prince A, Jac Fredo Agastinose Ronickom; Methodology: Gokul Manoj, Vaibhavi Gupta, Aditi Bhattacharya, Amalin Prince A, Jac Fredo Agastinose Ronickom; Formal analysis and investigation: Gokul Manoj, Vaibhavi Gupta Aditi Bhattacharya, Shaik Gadda Abdul Aleem, Dhanvi Vedantham, Amalin Prince A, Jac Fredo Agastinose Ronickom; Software: Gokul Manoj, Vaibhavi Gupta Aditi Bhattacharya, Shaik Gadda Abdul Aleem, Dhanvi Vedantham; Data Evaluation: Shaik Gadda Abdul Aleem, Dhanvi Vedantham; Writing—original draft preparation: Gokul Manoj, Vaibhavi Gupta Aditi Bhattacharya, Amalin Prince A, Jac Fredo Agastinose Ronickom.; Writing—review and editing: Gokul Manoj, Vaibhavi Gupta Aditi Bhattacharya, Amalin Prince A, Jac Fredo Agastinose Ronickom.; Visualization: Gokul Manoj, Vaibhavi Gupta Aditi Bhattacharya, Jac Fredo Agastinose Ronickom; Supervision: Amalin Prince A, Jac Fredo Agastinose Ronickom; Project administration: Amalin Prince A, Jac Fredo Agastinose Ronickom.
Corresponding author
Ethics declarations
Ethics approval
The ABIDE dataset used in this study has previously undergone ethical review and approval before its release for research purposes.
Consent to participate
This study is based on the analysis of the publicly available Abide dataset, and as such, no direct participant involvement or consent to participate is applicable.
Consent to publish
Since the ABIDE dataset used in this study is publicly available and has been shared for research purposes, no specific consent to publish is required.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Manoj, G., Gupta, V., Bhattacharya, A. et al. Diagnostic classification of autism spectrum disorder using sMRI improves with the morphological distance-related features compared to morphological features. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18817-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18817-5