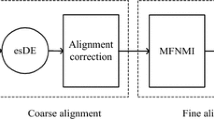
Abstract
In this paper, we develop an efficient mutual information based similarity metric for 3D medical image registration. The efficiency of the metric lies in the computation of mutual information, which uses modified algorithms for calculating entropy and joint entropy. We implemented the newly developed Efficient Entropy Mutual Information (EEMI) metric in SimpleITK, which is an open source image registration and segmentation toolkit. We designed 24 medical image registration frameworks using four similarity metrics, namely Mean Squares (MS), Joint Histogram Mutual Information (JHMI), Mattes Mutual Information (MattesMI) and our proposed EEMI, and six optimizers namely, Gradient Descent (GD), Conjugate Geadient Line Search (CGLS), 1+1 Evolutionary, Powell, Nelder-Mead (Amoeba) and Limited Memory Broyden Fletcher Goldfarb Shannon (LBFGS2). Using these frameworks, we performed elaborate comparative evaluation and analysis of EEMI in terms of registration accuracy and computation time. We used four different medical image data sets for our experiments. Data consisted of intra-modal and inter-modal image pairs of the brain and thorax obtained from different medical institutions as well as publicly available databases. Registration results prove the superiority and consistency of performance of EEMI compared to MS and JHMI. When compared with the benchmark MattesMI metric, performance of EEMI is on a par with respect to Dice score, Jaccard score and Hausdorff distance. With respect to computation time, EEMI is faster with GD, CGLS and Amoeba optimizers with 61.29%, 42.67% and 10.92% gain in computation times respectively. Among the other three optimizers, EEMI is more consistent with Powell’s optimizer. Mean, median and STD of TRE values of EEMI with Powell’s optimizer are respectively 67.42%, 65.84% and 74.63% less than that of MattesMI. Hausdorff distance of EEMI is 47.47% less than MattesMI, with Powell’s optimizer.












Similar content being viewed by others
Availability of Data
The data set analysed during the current study are available from the corresponding author on reasonable request.
References
Hajnal JV, Hill DLG, Hawkes DJ (eds) (2001) Medical image registration. The Biomedical Engineering Series, USA
Arun KS, Huang TS, Bostein SD (1987) Least-squares fitting of two 3D point sets. IEEE Trans Pattern Anal Mach Intell 9(5):698–700
Amit Y, Kong A (1996) Graphical templates for model registration. IEEE Trans Pattern Anal Mach Intell 18(3):225–236
Chua CS, Jarvis R (1996) 3D free-form surface registration and object recognition. Int J Comput Vision 17(1):77–99
Thirion JP (1996) New feature points based on geometric invariants for 3D image registration. Int J Comput Vision 18(2):121–137
Borgefors G (1986) Distance transformations in digital images. Comput Vision, Graph, Image Process 34:344–371
Jiang H, Robb RA, Holton KS (1992) New approach to 3-D registration of multi-modality medical images by surface matching. In: SPIE The international society for optical engineering. pp 196–196
Woods RP, Cherry SR, Mazziotta JC (1992) Rapid automated algorithm for aligning and reslicing PET images. J Comput Assist Tomograph 16(4):620–633
Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G (1995) Automated multi-modality image registration based on information theory. Inf Process Med Imaging 3(6):263–274
Viola P (1995) Alignment by maximization of mutual information. PhD thesis, MIT, Boston
Viola P, III WMW (1997) Alignment by maximization of mutual information. Int J Comput Vision 24(2):137–154
Cheng X, Zhang L, Zheng Y (2018) Deep similarity learning for multimodal medical images. Comput Methods Biomech Biomed Eng: Imaging Vis 6(3):248–252
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2019) Voxelmorph: A learning framework for deformable medical image registration. IEEE Trans Med Imaging 38(8):1788–1800
Lara-Hernández A, Rienmüller T, Juárez I, Pérez M, Reyna F, Baumgartner D, Makarenko VN, Bockeria OL, Maksudov M, Rienmüller R, Baumgartner C (2022) Deep learning-based image registration in dynamic myocardial perfusion CT imaging. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2022.3214380
Liu K, Ren Z, Wu X, Di J, Zhao J (2023) Ssg-net: A robust network for adaptive multi-source image registration based on superglue. Digit Signal Process 140
Krishnaswamy D, Noga M, Becher H, Boulanger P, Punithakumar K (2023) A novel 3D-to-3D Diffeomorphic registration algorithm with applications to left ventricle segmentation in MR and Ultrasound sequences. IEEE Access. 11:3144–3159
Zhang S, Shen J, Zheng S, Tang J (2023) Effective image registration model using optimized KAZE algorithm. Multimedia Tools Appl 1–26
Bouhlel F, Mliki H, Hammami M (2023) MOD-IR: moving objects detection from UAV-captured video sequences based on image registration. Multimedia Tools Appl 1–20
Deshmukh V, Khaparde A (2023) Depth map estimation with 3DFFT for two-dimensional to three-dimensional stereoscopic conversion based on image registration. Multimedia Tools Appl 1–28
Roy S, Bhattacharyya D, Bandyopadhyay SK, Kim TH (2017) An effective method for computerized prediction and segmentation of multiple sclerosis lesions in brain MRI. Comput Methods Program Biomed 140:307–320
Roy S, Bandyopadhyay SK (2016) A new method of brain tissues segmentation from MRI with accuracy estimation. Procedia Comput Sci 85:362–369
Roy S, Bhattacharyya D, Bandyopadhyay SK, Kim TH (2017) An iterative implementation of level set for precise segmentation of brain tissues and abnormality detection from MR images. IETE J Res 63(6):769–783
Sengupta D, Gupta P, Biswas A (2020) An efficient method for computation of entropy and joint entropy of images. In: International conference on intelligent computing. pp 282–290. https://doi.org/10.1007/978-3-030-60799-9_24
SimpleITK https://simpleitk.org
ITK https://itk.org
Vandemeulebroucke J, Sarrut D, Clarysse P (2007) The POPI-model, a point-validated pixel-based breathing thorax model. In: Proc. XVth international conference on the use of computers in radiation therapy (ICCR)
Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W (2003) PET-CT image registration in the chest using free-form deformations. IEEE Trans Med Imaging 22(1):120–128
Shannon CE (1948) A mathematical theory of communication. The Bell Syst Tech J 27(3):379–423
Shannon CE (1949) Communication in the presence of noise. Proc IRE 37(1):10–21
Collignon A, Vandermeulen D, P.Suetens Marchal G (1995) 3D multi-modality medical image registration using feature space clustering. In: International conference on computer vision, virtual reality, and robotics in medicine. pp 195–204
3D-Slicer https://www.slicer.org
M.Holden Hill DL, Denton ER, Jarosz JM, Cox TC, Rohlfing T, Goodey J, Hawkes DJ (2000) Voxel similarity measures for 3D serial MR brain image registration. IEEE Trans Med Imaging 9(2):94–102
Nocedal J, Wright SJ (eds) (1999) Numerical Optimization. Springer, USA
Press WH, Teukolsky SA, Vetterling WT, Flannery BP (eds) (2002) Numerical Recipes In C. Cambridge University Press, Cambridge
Juels A, Wattenberg M (1995) Hillclimbing as a baseline method for the evaluation of stochastic optimization algorithms. Adv Neural Inf Process 8:430–436
Rudolf G (1997) Convergence properties of evolutionary algorithms. PhD thesis, Verlag Dr. Kovac, Humburg
Nelder JA, Mead R (1965) A simplex method for function minimization. The Comput J 7(4):308–313
Acknowledgements
We are thankful to the Department of Nuclear Medicine, AIIMS Delhi, India, Panacea Hospital, Kanpur India and Hi-Care Diagnostics, Kanpur, India for providing data for our work and for assisting us with their expertise. We are also thankful to the Department of Radiology, SSKM Kolkata, India for helping us in testing the work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sengupta, D., Gupta, P. & Biswas, A. An efficient similarity metric for 3D medical image registration. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18710-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18710-1