Abstract
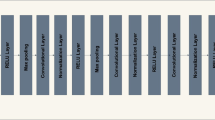
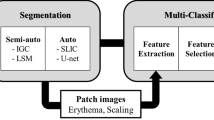
Psoriasis Severity and Area Index (PASI) is a gold standard scoring system for the assessment of Psoriasis skin disease. Generally, PASI scoring is done manually by expert dermatologists through visual and touch senses for psoriasis diagnosis and their treatment’s validation. This subjective approach raises several limitations and becomes unreliable. Many conventional and machine learning-based works are proposed for objective estimation of psoriasis area and severity from 2D RGB images. However, these works are validated on small datasets, require manual pre-processing, and rely heavily on hand-crafted features. In the proposed work, a fully automated system based on deep learning is designed for automated PASI scoring from raw 2D RGB images. This system contains a segmentation and three classification models for objective estimation of psoriasis area and severity scores for all three clinical symptoms of psoriasis, respectively. The psoriasis area is estimated by segmenting healthy and unhealthy regions simultaneously using a lightweight network as a backbone with UNet. After segmentation, the severity scores for each segmented lesion are automatically estimated by using a hybrid classification model. This model is developed by adopting a lightweight network for local feature extraction and integrating it with a vision transformer for learning global features. The psoriasis dataset used in the proposed work is self-prepared and contains 1,018 photographic images from different body regions of 212 psoriasis patients. The exhaustive performance analysis is done for the automatic estimation of each parameter of PASI. The proposed work achieves mean absolute error of 0.04, 0.23, 0.22, and 0.21 for objective estimation of Area, Redness, Scaliness, and Thickness scores, respectively. The mean absolute error obtained by the proposed system for automatic scoring of PASI is 1.02. The comparative studies with existing works further validate the efficacy of the proposed work. This work can further be improvised by using data from multi-centre and regions in a large population.







Similar content being viewed by others
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.
References
Shrivastava VK, Londhe ND, Sonawane RS, Suri JS (2015) First review on psoriasis severity risk stratification: an engineering perspective. Comput Biol Med 63:52–63
Nestle FO, Conrad C (2004) Mechanisms of psoriasis. Drug Discov Today: Dis Mech 1(3):315–319
Henseler T (1997) The genetics of psoriasis. J Am Acad Dermatol 37(2):S1–S11
Puzenat E, Bronsard V, Prey S, Gourraud PA, Aractingi S, Bagot M, ..., Aubin F (2010) What are the best outcome measures for assessing plaque psoriasis severity? A systematic review of the literature. J Eur Acad Dermatol Venereol 24:10–16
Chandran V, Raychaudhuri SP (2010) Geoepidemiology and environmental factors of psoriasis and psoriatic arthritis. J Autoimmun 34(3):J314–J321
Olivier C, Robert PD, Daihung DO, Urbà G, Catalin MP, Hywel W, ..., Gelfand JM (2010) The risk of depression, anxiety, and suicidality in patients with psoriasis: a population-based cohort study. Arch Dermatol 146(8):891–895
Huerta C, Rivero E, Rodríguez LAG (2007) Incidence and risk factors for psoriasis in the general population. Arch Dermatol 143(12):1559–1565
Menter A, Korman NJ, Elmets CA, Feldman SR, Gelfand JM, Gordon KB, Bhushan R (2011) Guidelines of care for the management of psoriasis and psoriatic arthritis: Sect. 6. Guidelines of care for the treatment of psoriasis and psoriatic arthritis: case-based presentations and evidence-based conclusions. J Am Acad Dermatol 65(1):137–174
Schmitt J, Wozel G (2005) The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasis. Dermatology 210(3):194–199
Feldman SR, Krueger G (2005) Psoriasis assessment tools in clinical trials. Ann Rheum Dis 64(suppl 2):ii65–ii68
Fink C, Alt C, Uhlmann L, Klose C, Enk A, Haenssle HA (2018) Intra-and interobserver variability of image‐based PASI assessments in 120 patients suffering from plaque‐type psoriasis. J Eur Acad Dermatol Venereol 32(8):1314–1319
Chalmers RJ (2015) Assessing psoriasis severity and outcomes for clinical trials and routine clinical practice. Dermatol Clin 33(1):57–71
Maglogiannis I, Doukas CN (2009) Overview of advanced computer vision systems for skin lesions characterization. IEEE Trans Inf Technol Biomed 13(5):721–733
Chang W-Y, Huang A, Yang C-Y, Lee C-H, Chen Y-C, Wu T-Y (2013) Computer-aided diagnosis of skin lesions using conventional digital photography: a reliability and feasibility study. PLoS ONE 8(11):e76212
Razmjooy N, Somayeh Mousavi B, Soleymani F, Hosseini Khotbesara M (2013) A computer-aided diagnosis system for malignant melanomas. Neural Comput Appl 23:7–8
Dash M, Londhe ND, Ghosh S, Raj R, Sonawane RS (2020) A cascaded deep convolution neural network based CADx system for psoriasis lesion segmentation and severity assessment. Appl Soft Comput 91:106240
Morrow T (2004) Evaluating new therapies for psoriasis. Manag Care 13:34–40
Balestrieri E, Lamonaca F, Lembo S, Miele G, Cusano F, De Cristofaro GA (2019) Automatic psoriasis assessment methods: current scenario and perspectives from a metrologic point of view. In: 2019 IEEE International Symposium on Medical Measurements and Applications (MeMeA), IEEE, pp. 1–6
Yu K, Syed MN, Bernardis E, Gelfand JM (2020) Machine learning applications in the evaluation and management of psoriasis: a systematic review. J Psoriasis Psoriatic Arthritis 5(4):147–159
Lu J, Kazmiercazk E, Manton JH, Sinclair R (2012) Automatic scoring of erythema and scaling severity in psoriasis diagnosis. In: AI 2012: Advances in Artificial Intelligence: 25th Australasian Joint Conference, Sydney, Australia, December 4–7, 2012. Proceedings 25 (pp. 73–84). Springer Berlin Heidelberg
Banu S, Toacse G, Danciu G (2014) Objective erythema assessment of Psoriasis lesions for Psoriasis Area and Severity Index (PASI) evaluation. In: 2014 International Conference and Exposition on Electrical and Power Engineering (EPE), IEEE, pp. 052–056
Raina A, Hennessy R, Rains M, Allred J, Hirshburg JM, Diven DG, Markey MK (2016) Objective measurement of erythema in psoriasis using digital color photography with color calibration. Skin Res Technol 22(3):375–380
George Y, Aldeen M, Garnavi R (2018) Psoriasis image representation using patch-based dictionary learning for erythema severity scoring. Comput Med Imaging Graph 66:44–55
George Y, Aldeen M, Garnavi R (2019) Automatic scale severity assessment method in psoriasis skin images using local descriptors. IEEE J Biomedical Health Inf 24(2):577–585
Serte S, Serener A, Al-Turjman F (2022) Deep learning in medical imaging: a brief review. Trans Emerg Telecommun Technol 33(10):e4080
Li LF, Wang X, Hu WJ, Xiong NN, Du YX, Li BS (2020) Deep learning in skin disease image recognition: a review. IEEE Access 8:208264–208280
Mathew A, Amudha P, Sivakumari S (2021) Deep learning techniques: an overview. Adv Mach Learn Technol Appl: Proc AMLTA 2020:599–608
Pal A, Chaturvedi A, Garain U, Chandra A, Chatterjee R (2016) Severity grading of psoriatic plaques using deep CNN based multi-task learning. In: 2016 23rd International Conference on Pattern Recognition (ICPR), IEEE, pp. 1478–1483
Pal A, Chaturvedi A, Garain U, Chandra A, Chatterjee R, Senapati S (2018) Severity assessment of psoriatic plaques using deep cnn based ordinal classification. In: OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis: First International Workshop, OR 2.0 2018, 5th International Workshop, CARE 2018, 7th International Workshop, CLIP 2018, Third International Workshop, ISIC 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16 and 20, 2018, Proceedings 5 (pp. 252–259). Springer International Publishing
Tancharoen D, Tantawiwat P, Kovintavewat P (2019) Medical imaging using automatic region of interest segmentation for psoriasis diagnosis. In: 2019 34th International Technical Conference on Circuits/Systems, Computers and Communications (ITC-CSCC), IEEE, pp. 1–4
Raj R, Londhe ND, Sonawane RS (2021) Deep learning based multi-segmentation for automatic estimation of psoriasis area score. In: 2021 8th International Conference on Signal Processing and Integrated Networks (SPIN), IEEE pp. 1137–1142
Fink C, Fuchs T, Enk A, Haenssle HA (2018) Design of an algorithm for automated, computer-guided PASI measurements by digital image analysis. J Med Syst 42:1–8
Li Y, Wu Z, Zhao S, Wu X, Kuang Y, Yan Y, …, Wang Y (2020) PSENet:Psoriasis severity evaluation network. In: Proceedings of the AAAI Conference on Artificial Intelligence 34(01):800–807
Wu X, Yan Y, Zhao S, Kuang Y, Ge S, Wang K, Chen X (2021) Automatic severity rating for improved psoriasis treatment. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VII 24 (pp. 185–194). Springer International Publishing
Schaap MJ, Cardozo NJ, Patel A, De Jong EMGJ, Van Ginneken B, Seyger MMB (2022) Image-based automated psoriasis area severity index scoring by convolutional neural networks. J Eur Acad Dermatol Venereol 36(1):68–75
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520
Vaswani A, Shazeer N, Parmar N, Uszkoreit, J, Jones L, Gomez AN, …, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst 30
Mehta S, Rastegari M (2022) MobileViT: light-weight, general-purpose, and mobile-friendly vision transformer. Computer Vision and Pattern Recognition. https://doi.org/10.48550/arXiv.2110.02178
Fernández A, García S, Galar M, Prati RC, Krawczyk B, Herrera F, …, Herrera F (2018) Cost-sensitive learning. Learning from Imbalanced Data Sets 63–78
Johnson JM, Khoshgoftaar TM (2019) Survey on deep learning with class imbalance. J Big Data 6(1):1–54
Song B, Li S, Sunny S, Gurushanth K, Mendonca P, Mukhia N, …, Liang R (2021) Classification of imbalanced oral cancer image data from high-risk population. J Biomed Optics 26(10):105001–105001
Zhuang F, Qi Z, Duan K, Xi D, Zhu Y, Zhu H, …, He Q (2020) A comprehensive survey on transfer learning. Proc IEEE 109(1):43–76
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J big data 6(1):1–48
Anaya-Isaza A, Mera-Jiménez L (2022) Data augmentation and transfer learning for brain tumor detection in magnetic resonance imaging. IEEE Access 10:23217–23233
Rai R, Sisodia DS (2021) Real-time data augmentation based transfer learning model for breast cancer diagnosis using histopathological images. In: Advances in Biomedical Engineering and Technology: Select Proceedings of ICBEST 2018 (pp. 473–488). Springer Singapore
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18 (pp. 234–241). Springer International Publishing
Albawi S, Mohammed TA, Al-Zawi S (2017) Understanding of a convolutional neural network. In: 2017 International Conference on Engineering and Technology (ICET), IEEE, pp. 1–6
Han K, Wang Y, Chen H, Chen X, Guo J, Liu Z, …, Tao D (2022) A survey on vision transformer. IEEE Trans Pattern Anal Mach Intell 45(1):87–110
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, …, Fei-Fei L (2015) Imagenet large scale visual recognition challenge. Int J Comput Vis 115:211–252
Xiao T, Singh M, Mintun E, Darrell T, Dollár P, Girshick R (2021) Early convolutions help transformers see better. Adv Neural Inf Process Syst 34:30392–30400
Koffas S, Picek S, Conti M (2022) Dynamic backdoors with global average pooling. In: 2022 IEEE 4th International Conference on Artificial Intelligence Circuits and Systems (AICAS), IEEE, pp. 320–323
Kumar RL, Kakarla J, Isunuri BV, Singh M (2021) Multi-class brain tumor classification using residual network and global average pooling. Multimed Tools Appl 80:13429–13438
Errichetti E, Stinco G (2016) Dermoscopy in general dermatology: a practical overview. Dermatol Ther 6:471–507
Anand V, Gupta S, Nayak SR, Koundal D, Prakash D, Verma KD (2022) An automated deep learning models for classification of skin disease using dermoscopy images: a comprehensive study. Multimed Tools Appl 81(26):37379–37401
Lei J (2020) Cross-validation with confidence. J Am Stat Assoc 115(532):1978–1997
Python W (2021) Python. Python releases for windows, 24
Chollet F (2018) Keras: the python deep learning library. Astrophysics source code library, pp ascl–1806
Martín A, Ashish A, Paul B, Eugene B, Zhifeng C, Craig C, …, Matthieu D (2015) TensorFlow: Large-scale machine learning on heterogeneous systems. Software available from tensorflow.org
King G, Zeng L (2001) Logistic regression in rate events data, Harvard University. Center for Basic Research in the Social Sciences
Koidl K (2013) Loss functions in classification tasks. School of Computer Science and Statistic Trinity College, Dublin
Kingma DP, Ba J (2015) Adam: A method for stochastic optimization, 3rd International Conference for Learning Representations, San Diego. https://doi.org/10.48550/arXiv.1412.6980
Setiawan AW (2020) Image segmentation metrics in skin lesion: accuracy, sensitivity, specificity, dice coefficient, Jaccard index, and Matthews correlation coefficient. In: 2020 International Conference on Computer Engineering, Network, and Intelligent Multimedia (CENIM), IEEE, pp. 97–102
Grandini M, Bagli E, Visani G (2020) Metrics for multi-class classification: an overview. arXiv preprint arXiv:2008.05756
Mortaz E (2020) Imbalance accuracy metric for model selection in multi-class imbalance classification problems. Knowl Based Syst 210:106490
Hoo ZH, Candlish J, Teare D (2017) What is an ROC curve? Emerg Med J 34(6):357–359
Narkhede S (2018) Understanding auc-roc curve. Towards Data Science 26(1):220–227
Willmott CJ, Matsuura K (2005) Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance. Climate Res 30(1):79–82
Chai T, Draxler RR (2014) Root mean square error (RMSE) or mean absolute error (MAE)?–Arguments against avoiding RMSE in the literature. Geosci Model Dev 7(3):1247–1250
Bartko JJ (1966) The intraclass correlation coefficient as a measure of reliability. Psychol Rep 19(1):3–11
Krstinić D, Braović M, Šerić L, Božić-Štulić D (2020) Multi-label classifier performance evaluation with confusion matrix. Computer Science & Information Technology 1. https://doi.org/10.5121/csit.2020.100801
Zivkovic M, Bacanin N, Antonijevic M, Nikolic B, Kvascev G, Marjanovic M, Savanovic N (2022) Hybrid CNN and XGBoost model tuned by modified arithmetic optimization algorithm for COVID-19 early diagnostics from X-ray images. Electronics 11(22):3798
Acknowledgements
We thank all the dermatologists and psoriasis patients of Psoriasis Clinic and Research Centre, Psoriatreat, Pune, Maharashtra, India who are involved in this research.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation was performed by Ritesh Raj and Narendra Londhe. Data collection and analysis were performed by Ritesh Raj and Rajendra Sonawane. The first draft of the manuscript was written by Ritesh Raj and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Raj, R., Londhe, N.D. & Sonawane, R.S. Objective scoring of psoriasis area and severity index in 2D RGB images using deep learning. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18138-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18138-7